













| General Information: |
|
| Name(s) found: |
LAS1L
[HGNC (HUGO)]
|
| Description(s) found:
Found 37 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 734 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSWESGAGPG LGSQGMDLVW SAWYGKCVKG KGSLPLSAHG IVVAWLSRAE WDQVTVYLFC 60
61 DDHKLQRYAL NRITVWRSRS GNELPLAVAS TADLIRCKLL DVTGGLGTDE LRLLYGMALV 120
121 RFVNLISERK TKFAKVPLKC LAQEVNIPDW IVDLRHELTH KKMPHINDCR RGCYFVLDWL 180
181 QKTYWCRQLE NSLRETWELE EFREGIEEED QEEDKNIVVD DITEQKPEPQ DDGKSTESDV 240
241 KADGDSKGSE EVDSHCKKAL SHKELYERAR ELLVSYEEEQ FTVLEKFRYL PKAIKAWNNP 300
301 SPRVECVLAE LKGVTCENRE AVLDAFLDDG FLVPTFEQLA ALQIEYEDGQ TEVQRGEGTD 360
361 PKSHKNVDLN DVLVPKPFSQ FWQPLLRGLH SQNFTQALLE RMLSELPALG ISGIRPTYIL 420
421 RWTVELIVAN TKTGRNARRF SAGQWEARRG WRLFNCSASL DWPRMVESCL GSPCWASPQL 480
481 LRIIFKAMGQ GLPDEEQEKL LRICSIYTQS GENSLVQEGS EASPIGKSPY TLDSLYWSVK 540
541 PASSSFGSEA KAQQQEEQGS VNDVKEEEKE EKEVLPDQVE EEEENDDQEE EEEDEDDEDD 600
601 EEEDRMEVGP FSTGQESPTA ENARLLAQKR GALQGSAWQV SSEDVRWDTF PLGRMPGQTE 660
661 DPAELMLENY DTMYLLDQPV LEQRLEPSTC KTDTLGLSCG VGSGNCSNSS SSNFEGLLWS 720
721 QGQLHGLKTG LQLF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample pIC145 from april 2005 | Okada, M., et al. (2006) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
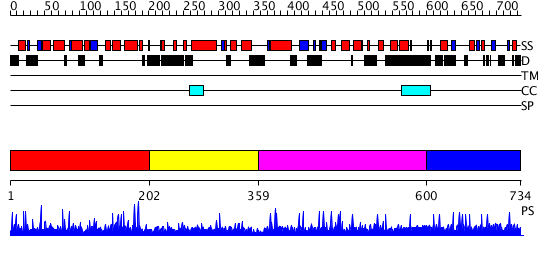
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..201] | 93.236572 | No description for PF04031.2 was found. No confident structure predictions are available. | |
| 2 | View Details | [202..358] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [359..599] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [600..734] | 1.002938 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)