













| General Information: |
|
| Name(s) found: |
EXOSC10
[HGNC (HUGO)]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Homo sapiens |
| Length: | 885 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPPSTREPR VLSATSATKS DGEMVLPGFP DADSFVKFAL GSVVAVTKAS GGLPQFGDEY 60
61 DFYRSFPGFQ AFCETQGDRL LQCMSRVMQY HGCRSNIKDR SKVTELEDKF DLLVDANDVI 120
121 LERVGILLDE ASGVNKNQQP VLPAGLQVPK TVVSSWNRKA AEYGKKAKSE TFRLLHAKNI 180
181 IRPQLKFREK IDNSNTPFLP KIFIKPNAQK PLPQALSKER RERPQDRPED LDVPPALADF 240
241 IHQQRTQQVE QDMFAHPYQY ELNHFTPADA VLQKPQPQLY RPIEETPCHF ISSLDELVEL 300
301 NEKLLNCQEF AVDLEHHSYR SFLGLTCLMQ ISTRTEDFII DTLELRSDMY ILNESLTDPA 360
361 IVKVFHGADS DIEWLQKDFG LYVVNMFDTH QAARLLNLGR HSLDHLLKLY CNVDSNKQYQ 420
421 LADWRIRPLP EEMLSYARDD THYLLYIYDK MRLEMWERGN GQPVQLQVVW QRSRDICLKK 480
481 FIKPIFTDES YLELYRKQKK HLNTQQLTAF QLLFAWRDKT ARREDESYGY VLPNHMMLKI 540
541 AEELPKEPQG IIACCNPVPP LVRQQINEMH LLIQQAREMP LLKSEVAAGV KKSGPLPSAE 600
601 RLENVLFGPH DCSHAPPDGY PIIPTSGSVP VQKQASLFPD EKEDNLLGTT CLIATAVITL 660
661 FNEPSAEDSK KGPLTVAQKK AQNIMESFEN PFRMFLPSLG HRAPVSQAAK FDPSTKIYEI 720
721 SNRWKLAQVQ VQKDSKEAVK KKAAEQTAAR EQAKEACKAA AEQAISVRQQ VVLENAAKKR 780
781 ERATSDPRTT EQKQEKKRLK ISKKPKDPEP PEKEFTPYDY SQSDFKAFAG NSKSKVSSQF 840
841 DPNKQTPSGK KCIAAKKIKQ SVGNKSMSFP TGKSDRGFRY NWPQR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
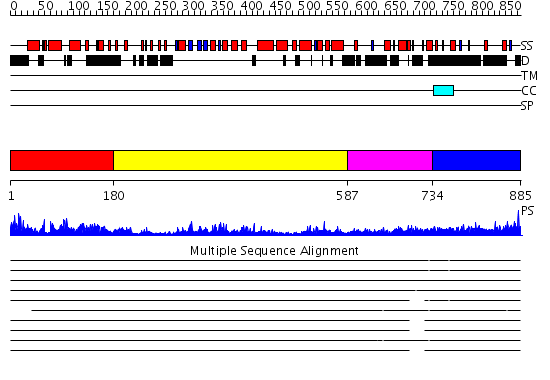
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..179] | 24.69897 | Divalent metal ions (manganese) bound to T5 5'-exonuclease | |
| 2 | View Details | [180..586] | 53.30103 | No description for 2hbjA was found. | |
| 3 | View Details | [587..733] | 50.69897 | Exonuclease domain of prokaryotic DNA polymerase; DNA polymerase I (Klenow fragment) | |
| 4 | View Details | [734..885] | 12.260997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)