













| General Information: |
|
| Name(s) found: |
ROC2_ORYSJ
[Swiss-Prot]
|
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 784 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMIPARHMPS MIGRNGAAYG SSSALSLSQP NLLDNHQFQQ AFQHQQQQHH LLDQIPATTA 60
61 ESGDNMIRSR ASDPLGGDEF ESKSGSENVD GVSVDDQDPN QRPRKKRYHR HTQHQIQEME 120
121 AFFKECPHPD DKQRKELSRE LGLEPLQVKF WFQNKRTQMK NQHERHENSQ LRSDNEKLRA 180
181 ENMRYKEALS SASCPNCGGP AALGEMSFDE HHLRIENARL REEIDRISAI AAKYVGKPMV 240
241 PFPVLSNPMA AAASRAPLDL PVAPYGVPGD MFGGGGAGEL LRGVQSEVDK PMIVELAVAA 300
301 MEELVRMAQL DEPLWSVAPP LDATAAAMET LSEEEYARMF PRGLGPKQYG LRSEASRDSA 360
361 VVIMTHANLV EILMDANQYA AVFSNIVSRA ITLEVLSTGV AGNYNGALQV MSVEFQVPSP 420
421 LVPTRESYFV RYCKQNADGT WAVVDVSLDS LRPSPVLKCR RRPSGCLIQE MPNGYSKVTW 480
481 VEHVEVDDRS VHNIYKLLVN SGLAFGARRW VGTLDRQCER LASVMASNIP TSDIGVITSS 540
541 EGRKSMLKLA ERMVVSFCGG VTASVAHQWT TLSGSGAEDV RVMTRKSVDD PGRPPGIVLN 600
601 AATSFWLPVP PKRVFDFLRD ESSRSEWDIL SNGGIVQEMA HIANGRDQGN CVSLLRVNSS 660
661 NSNQSNMLIL QESCTDASGS YVIYAPVDVV AMNVVLNGGD PDYVALLPSG FAILPDGPAH 720
721 DGGDGDGGVG VGSGGSLLTV AFQILVDSVP TAKLSLGSVA TVNSLIACTV ERIKAAVSGE 780
781 SNPQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
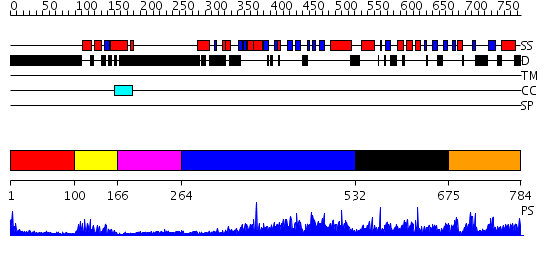
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [100..165] | 19.69897 | Paired protein | |
| 3 | View Details | [166..263] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [264..531] | 2.51 | No description for 2r55A was found. | |
| 5 | View Details | [532..674] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [675..784] | 3.065979 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)