













| General Information: |
|
| Name(s) found: |
RACGAP1
[HGNC (HUGO)]
|
| Description(s) found:
Found 39 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 632 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTMMLNVRN LFEQLVRRVE ILSEGNEVQF IQLAKDFEDF RKKWQRTDHE LGKYKDLLMK 60
61 AETERSALDV KLKHARNQVD VEIKRRQRAE ADCEKLERQI QLIREMLMCD TSGSIQLSEE 120
121 QKSALAFLNR GQPSSSNAGN KRLSTIDESG SILSDISFDK TDESLDWDSS LVKTFKLKKR 180
181 EKRRSTSRQF VDGPPGPVKK TRSIGSAVDQ GNESIVAKTT VTVPNDGGPI EAVSTIETVP 240
241 YWTRSRRKTG TLQPWNSDST LNSRQLEPRT ETDSVGTPQS NGGMRLHDFV SKTVIKPESC 300
301 VPCGKRIKFG KLSLKCRDCR VVSHPECRDR CPLPCIPTLI GTPVKIGEGM LADFVSQTSP 360
361 MIPSIVVHCV NEIEQRGLTE TGLYRISGCD RTVKELKEKF LRVKTVPLLS KVDDIHAICS 420
421 LLKDFLRNLK EPLLTFRLNR AFMEAAEITD EDNSIAAMYQ AVGELPQANR DTLAFLMIHL 480
481 QRVAQSPHTK MDVANLAKVF GPTIVAHAVP NPDPVTMLQD IKRQPKVVER LLSLPLEYWS 540
541 QFMMVEQENI DPLHVIENSN AFSTPQTPDI KVSLLGPVTT PEHQLLKTPS SSSLSQRVRS 600
601 TLTKNTPRFG SKSKSATNLG RQGNFFASPM LK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
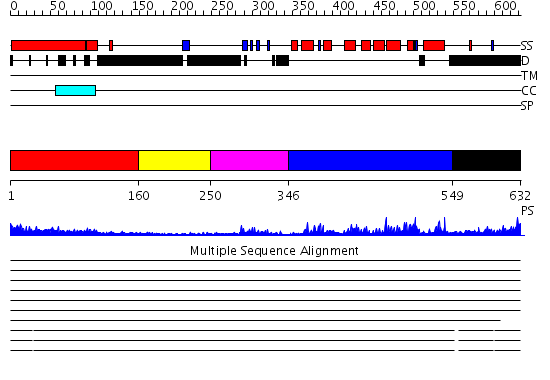
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..159] | 7.69897 | Heavy meromyosin subfragment | |
| 2 | View Details | [160..249] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [250..345] | 3.69897 | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE | |
| 4 | View Details | [346..548] | 42.09691 | No description for 2ovjA was found. | |
| 5 | View Details | [549..632] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)