













| General Information: |
|
| Name(s) found: |
egl-9 /
CE40254
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 647 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein metabolic process
[IEA |
| Molecular Function: |
zinc ion binding
[IEA oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MICKSLQTSG MVPSNLMPQA APAVMAPIPP TVSFDDPALT TSLLLSLQNN PILNQTISNF 60
61 PPTFSITSKT EPEPSIPIQI PQRISSTSTV PFSSEGSAFK PYRNTHVFNS ISSESMSSMC 120
121 TSHEASLEHM SSASLAMFPT SSTAQSDISR LAQVLSLAGD SPASLALVTT SVPSTASTAT 180
181 IPPPATTTSS ATSSGKSETI TVGKEKIIQT DDPDIQIIET EGGSKPTVSR TRKRPTPSNS 240
241 ADPKINYKDH NKNVVYSTTL QEHQKHLQNR GLALSIHQAM VLRLRYIAEH VIRSLNEFGW 300
301 AVVDNFLGSD HYKFTAKEIE RLYERGLFSP GQLMEAKHKD EFHIKDIRSD HIYWYDGYDG 360
361 RAKDAATVRL LISMIDSVIQ HFKKRIDHDI GGRSRAMLAI YPGNGTRYVK HVDNPVKDGR 420
421 CITTIYYCNE NWDMATDGGT LRLYPETSMT PMDIDPRADR LVFFWSDRRN PHEVMPVFRH 480
481 RFAITIWYMD KSERDKALAK GKESDAACAS KKENDPTSSS LNSLIGSLLR PRKNPSTHDL 540
541 SKLDLRLFPS TSSDPALVSA ADEDRVDISA DFQSTSSLAH PESTDSGVSL STFNVAHNHM 600
601 ERTTSLQSIS DHFRSERSHE RRSSTSSDQD LDEGLPPPPS TNPEYYI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
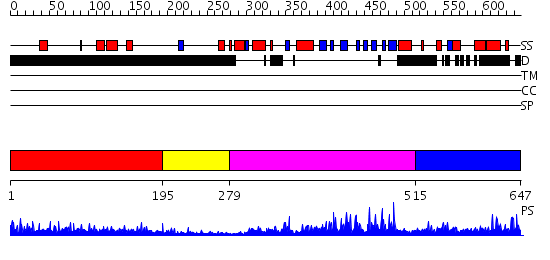
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..194] | 11.176997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [195..278] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [279..514] | 86.221849 | No description for 2hbtA was found. | |
| 4 | View Details | [515..647] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.75 |
Source: Reynolds et al. (2008)