













| General Information: |
|
| Name(s) found: |
FAB1A_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1757 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cellular protein metabolic process
[IEA]
phosphatidylinositol metabolic process [IEA] |
| Molecular Function: |
ATP binding
[IEA]
zinc ion binding [IEA][ISS] phosphatidylinositol phosphate kinase activity [IEA] protein binding [IEA] 1-phosphatidylinositol-4-phosphate 5-kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSQDHKAPG FVDIVKSWIP RKSESSNMSR DFWMPDQSCP VCYECDAQFT VFNRRHHCRL 60
61 CGRVFCAKCA ANSIPSPSDE TKDSHEEPER IRVCNYCYKQ WEQGIVPPDN GASIISLHFS 120
121 SSPSARSVAS TTSNSSNCTI DSTAGPSPRP KMNPRASRRV SSNMDSEKSE QQNAKSRRSS 180
181 DHYGHVLDSS DNQVEFFVNS SGRSDGEADD DDDYQSDFAQ SYAQGNDYYG AINLDEVDHI 240
241 YGSHEAHDVG VKIEPNISGF PPDQDLDSLN TETIDKTRQQ ENGWNDVKEG SPPCEESFEP 300
301 EVVDFESDGL LWLPPEPENE EDEREAVLSD DDGDEGDRGD WGYLRPSNSF NEKDFHSKDK 360
361 SSGAMKNVVE GHFRALVAQL LEVDNLPMVN EGDEEGWLDI ITSLSWEAAT LLKPDTSKSG 420
421 GMDPGGYVKV KCIPCGRRSE SMVVKGVVCK KNVAHRRMTS KIEKPRLLIL GGALEYQRIS 480
481 NQLSSFDTLL QQEMDHLKMA VAKIDSHNPD ILLVEKSVSR FAQEYLLAKD ISLVLNIKRS 540
541 LLERISRCTG AQIVPSIDQL TSPKLGYCDL FHVEKFVETH VSPCQVAKKM AKTLMFFDGC 600
601 PKPLGCTILL KGAHEDELKK VKHVIQYGVF AAYHLALETS FLADEGASIH ELPLQTPITV 660
661 ALPDKPSMVN RSISTIPGFT VSSAEKSPTT ELRGEPHKAN GDLTGNFTSS KTHFQGKLDG 720
721 NDRIDPSERL LHNLDTVYCK PPETITSKDD GLVPTLESRQ LSFHVEEPSV QKDQWSVLSG 780
781 ATEQVTDGGY TNDSAVIGNQ NFNRQEQMES SKGDFHPSAS DHQSILVSLS TRCVWKGSVC 840
841 ERAHLLRIKY YGSFDKPLGR FLRDNLFDQD QCCPSCTMPA EAHIHCYTHR QGSLTISVKK 900
901 LPELLPGQRE GKIWMWHRCL KCPRINGFPP ATRRIVMSDA AWGLSFGKFL ELSFSNHAAA 960
961 SRVANCGHSL HRDCLRFYGF GRMVACFRYA SINIYAVTLP PAKLYFNYEN QEWLQKESKE1020
1021 VIKKAEVLFN EVQEALSQIS AKTMGAGSKG STPNKIKLSL EELAGLLEQR KKEYKDSLQQ1080
1081 MLNVVKDGQP TIDILLINKL RRLIIFDSYA WDECLAGAAN MVRNNYLEAP KNSAPKVMGR1140
1141 NVSLEKLSDE KVKSIPTHVA ICNDSLLQDA DYETCLNQGK SFADTSGKFA IPEDVGSDRP1200
1201 PDCRMEFDPS EGGKDNFVES SQVVKPAHTE SQFQATDLSD TLDAAWIGEQ TTSENGIFRP1260
1261 PSRAASTNGT QIPDLRLLGS ESELNFKGGP TNDEHTTQVQ LPSPSFYYSL NKNYSLNSRK1320
1321 HIMAEDRPVY VSSYRELEWR SGARLLLPLG CNDLVLPVYD DEPTSIIAYA LTSSEYKAQM1380
1381 SGSDKSRDRL DSGGSFSLFD SVNLLSLNSL SDLSVDMSRS LSSADEQVSQ LLHSSLYLKD1440
1441 LHARISFTDE GPPGKVKYSV TCYYAKEFEA LRMICCPSET DFIRSLGRCR KWGAQGGKSN1500
1501 VFFAKSLDDR FIIKQVTKTE LESFIKFGPA YFKYLTESIS TKSPTSLAKI LGIYQVSSKH1560
1561 LKGGKEFKMD VLVMENLLFK RNFTRLYDLK GSTRARYNPD TSGSNTVLLD QNLVEAMPTS1620
1621 PIFVGSKAKR LLERAVWNDT SFLASIHVMD YSLLVGVDEE RNELVLGIID FMRQYTWDKH1680
1681 LETWVKTSGL LGGPKNSTPT VISPQQYKKR FRKAMTAYFL MVPDQWSPAA VVPSNSSSAE1740
1741 VKEEEEKDNP QAVGNKS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
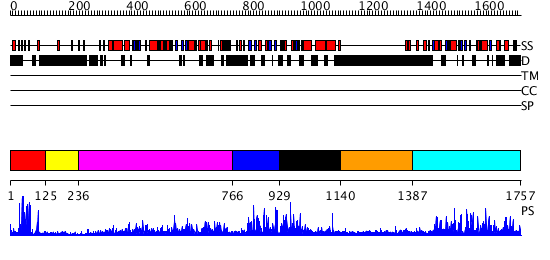
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..124] | 18.39794 | Eea1; Eea1 homodimerisation domain | |
| 2 | View Details | [125..235] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [236..765] | 116.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) | |
| 4 | View Details | [766..928] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [929..1139] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1140..1386] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1387..1757] | 79.30103 | Phosphatidylinositol phosphate kinase IIbeta, PIPK IIbeta |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)