













| General Information: |
|
| Name(s) found: |
gi|145332787
[NCBI NR]
gi|110738909 [NCBI NR] |
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 697 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plastid chromosome
[IDA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
RNA binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQICQTKLNF TFPNPTNPNF CKPKALQWSP PRRISLLPCR GFSSDEFPVD ETFLEKFGPK 60
61 DKDTEDEARR RNWIERGWAP WEEILTPEAD FARKSLNEGE EVPLQSPEAI EAFKMLRPSY 120
121 RKKKIKEMGI TEDEWYAKQF EIRGDKPPPL ETSWAGPMVL RQIPPRDWPP RGWEVDRKEL 180
181 EFIREAHKLM AERVWLEDLD KDLRVGEDAT VDKMCLERFK VFLKQYKEWV EDNKDRLEEE 240
241 SYKLDQDFYP GRRKRGKDYE DGMYELPFYY PGMVCEGTVT TLHLYQGAFV DIGGVHEGWV 300
301 PIKGNDWFWI RHFIKVGMHV IVEITVILRH IFFFRIYKNL NGNCFILLFL LFFQAKRDPY 360
361 RFRFPLELRF VHPNIDHMIF NKFDFPPIFH RDGDTNPDEI RRDCGRPPEP RKDPGSKPEE 420
421 EGLLSDHPYV DKLWQIHVAE QMILGDYEAN PAKYEGKKLS ELSDDEDFDE QKDIEYGEAY 480
481 YKKTKLPKVI LKTSVKELDL EAALTERQHH NKLMMEAKAR GEGYKIDKLR RNIEMDEYDF 540
541 LHWRRSLEER EALLRDISSR QALGLPLEEP GRYKPGSFFG KDQYDPTSAL YQYDYWGEPK 600
601 NSEISKQERM KDAHNKSIVG KGNVWYDMSY DDAIKQTIEK RKEGSTLASQ EEETESEEEE 660
661 EDDDDFDDFD YSILSDESSI GYSEQQPLVN GTQVLTD |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
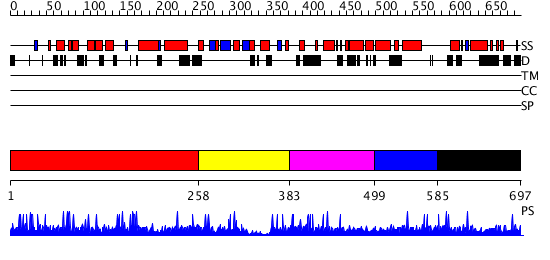
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..257] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [258..382] | 1.007973 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [383..498] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [499..584] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [585..697] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)