













| General Information: |
|
| Name(s) found: |
TSC1
[HGNC (HUGO)]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Homo sapiens |
| Length: | 1164 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQQANVGEL LAMLDSPMLG VRDDVTAVFK ENLNSDRGPM LVNTLVDYYL ETSSQPALHI 60
61 LTTLQEPHDK HLLDRINEYV GKAATRLSIL SLLGHVIRLQ PSWKHKLSQA PLLPSLLKCL 120
121 KMDTDVVVLT TGVLVLITML PMIPQSGKQH LLDFFDIFGR LSSWCLKKPG HVAEVYLVHL 180
181 HASVYALFHR LYGMYPCNFV SFLRSHYSMK ENLETFEEVV KPMMEHVRIH PELVTGSKDH 240
241 ELDPRRWKRL ETHDVVIECA KISLDPTEAS YEDGYSVSHQ ISARFPHRSA DVTTSPYADT 300
301 QNSYGCATST PYSTSRLMLL NMPGQLPQTL SSPSTRLITE PPQATLWSPS MVCGMTTPPT 360
361 SPGNVPPDLS HPYSKVFGTT AGGKGTPLGT PATSPPPAPL CHSDDYVHIS LPQATVTPPR 420
421 KEERMDSARP CLHRQHHLLN DRGSEEPPGS KGSVTLSDLP GFLGDLASEE DSIEKDKEEA 480
481 AISRELSEIT TAEAEPVVPR GGFDSPFYRD SLPGSQRKTH SAASSSQGAS VNPEPLHSSL 540
541 DKLGPDTPKQ AFTPIDLPCG SADESPAGDR ECQTSLETSI FTPSPCKIPP PTRVGFGSGQ 600
601 PPPYDHLFEV ALPKTAHHFV IRKTEELLKK AKGNTEEDGV PSTSPMEVLD RLIQQGADAH 660
661 SKELNKLPLP SKSVDWTHFG GSPPSDEIRT LRDQLLLLHN QLLYERFKRQ QHALRNRRLL 720
721 RKVIKAAALE EHNAAMKDQL KLQEKDIQMW KVSLQKEQAR YNQLQEQRDT MVTKLHSQIR 780
781 QLQHDREEFY NQSQELQTKL EDCRNMIAEL RIELKKANNK VCHTELLLSQ VSQKLSNSES 840
841 VQQQMEFLNR QLLVLGEVNE LYLEQLQNKH SDTTKEVEMM KAAYRKELEK NRSHVLQQTQ 900
901 RLDTSQKRIL ELESHLAKKD HLLLEQKKYL EDVKLQARGQ LQAAESRYEA QKRITQVFEL 960
961 EILDLYGRLE KDGLLKKLEE EKAEAAEAAE ERLDCCNDGC SDSMVGHNEE ASGHNGETKT1020
1021 PRPSSARGSS GSRGGGGSSS SSSELSTPEK PPHQRAGPFS SRWETTMGEA SASIPTTVGS1080
1081 LPSSKSFLGM KARELFRNKS ESQCDEDGMT SSLSESLKTE LGKDLGVEAK IPLNLDGPHP1140
1141 SPPTPDSVGQ LHIMDYNETH HEHS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
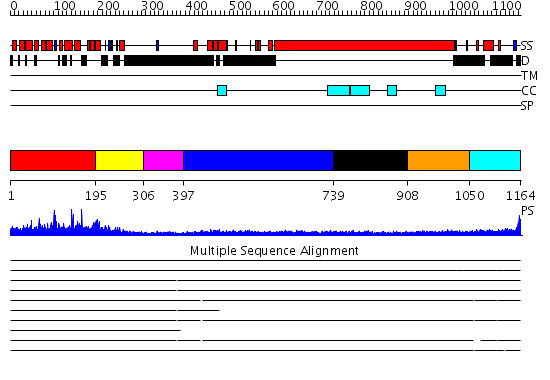
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..194] | 1.011947 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [195..305] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [306..396] | 6.522879 | Tropomyosin | |
| 4 | View Details | [397..738] | 24.0 | Heavy meromyosin subfragment | |
| 5 | View Details | [739..907] | 4.69897 | No description for 2i1jA was found. | |
| 6 | View Details | [908..1049] | 1.33 | Crystal structure of human GGA1 GAT domain complexed with the GAT-binding domain of Rabaptin5 | |
| 7 | View Details | [1050..1164] | 2.579996 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.50 |
Source: Reynolds et al. (2008)