













| General Information: |
|
| Name(s) found: |
DDX3Y
[HGNC (HUGO)]
|
| Description(s) found:
Found 39 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 660 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSHVVVKNDP ELDQQLANLD LNSEKQSGGA STASKGRYIP PHLRNREASK GFHDKDSSGW 60
61 SCSKDKDAYS SFGSRDSRGK PGYFSERGSG SRGRFDDRGR SDYDGIGNRE RPGFGRFERS 120
121 GHSRWCDKSV EDDWSKPLPP SERLEQELFS GGNTGINFEK YDDIPVEATG SNCPPHIENF 180
181 SDIDMGEIIM GNIELTRYTR PTPVQKHAIP IIKGKRDLMA CAQTGSGKTA AFLLPILSQI 240
241 YTDGPGEALK AVKENGRYGR RKQYPISLVL APTRELAVQI YEEARKFSYR SRVRPCVVYG 300
301 GADIGQQIRD LERGCHLLVA TPGRLVDMME RGKIGLDFCK YLVLDEADRM LDMGFEPQIR 360
361 RIVEQDTMPP KGVRHTMMFS ATFPKEIQML ARDFLDEYIF LAVGRVGSTS ENITQKVVWV 420
421 EDLDKRSFLL DILGATGSDS LTLVFVETKK GADSLEDFLY HEGYACTSIH GDRSQRDREE 480
481 ALHQFRSGKS PILVATAVAA RGLDISNVRH VINFDLPSDI EEYVHRIGRT GRVGNLGLAT 540
541 SFFNEKNMNI TKDLLDLLVE AKQEVPSWLE NMAYEHHYKG GSRGRSKSNR FSGGFGARDY 600
601 RQSSGSSSSG FGASRGSSSR SGGGGYGNSR GFGGGGYGGF YNSDGYGGNY NSQGVDWWGN 660
661 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample pIC146 from april 2005 | Okada, M., et al. (2006) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
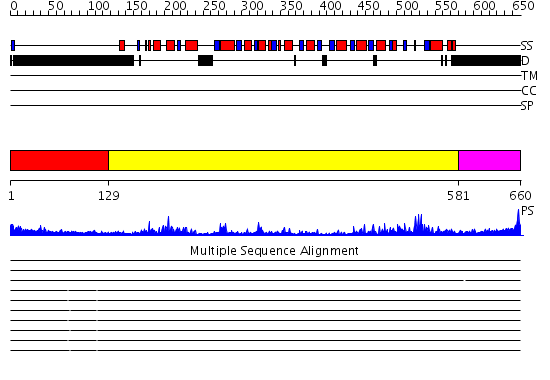
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [129..580] | 100.0 | No description for 2i4iA was found. | |
| 3 | View Details | [581..660] | 1.04 | Ribonuclease domain of colicin E3; Colicin E3 translocation domain; Colicin E3 receptor domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)