













| General Information: |
|
| Name(s) found: |
DNAJA4
[HGNC (HUGO)]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 397 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKETQYYDI LGVKPSASPE EIKKAYRKLA LKYHPDKNPD EGEKFKLISQ AYEVLSDPKK 60
61 RDVYDQGGEQ AIKEGGSGSP SFSSPMDIFD MFFGGGGRMA RERRGKNVVH QLSVTLEDLY 120
121 NGVTKKLALQ KNVICEKCEG VGGKKGSVEK CPLCKGRGMQ IHIQQIGPGM VQQIQTVCIE 180
181 CKGQGERINP KDRCESCSGA KVIREKKIIE VHVEKGMKDG QKILFHGEGD QEPELEPGDV 240
241 IIVLDQKDHS VFQRRGHDLI MKMKIQLSEA LCGFKKTIKT LDNRILVITS KAGEVIKHGD 300
301 LRCVRDEGMP IYKAPLEKGI LIIQFLVIFP EKHWLSLEKL PQLEALLPPR QKVRITDDMD 360
361 QVELKEFCPN EQNWRQHREA YEEDEDGPQA GVQCQTA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
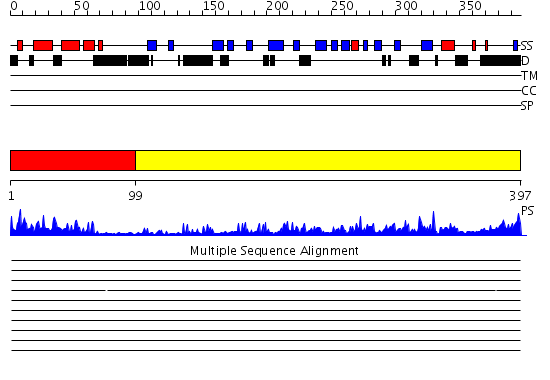
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | 22.30103 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [99..397] | 54.522879 | The crystal structure of Hsp40 Ydj1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)