













| General Information: |
|
| Name(s) found: |
GGA1
[HGNC (HUGO)]
|
| Description(s) found:
Found 40 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 639 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEPAMEPETL EARINRATNP LNKELDWASI NGFCEQLNED FEGPPLATRL LAHKIQSPQE 60
61 WEAIQALTVL ETCMKSCGKR FHDEVGKFRF LNELIKVVSP KYLGSRTSEK VKNKILELLY 120
121 SWTVGLPEEV KIAEAYQMLK KQGIVKSDPK LPDDTTFPLP PPRPKNVIFE DEEKSKMLAR 180
181 LLKSSHPEDL RAANKLIKEM VQEDQKRMEK ISKRVNAIEE VNNNVKLLTE MVMSHSQGGA 240
241 AAGSSEDLMK ELYQRCERMR PTLFRLASDT EDNDEALAEI LQANDNLTQV INLYKQLVRG 300
301 EEVNGDATAG SIPGSTSALL DLSGLDLPPA GTTYPAMPTR PGEQASPEQP SASVSLLDDE 360
361 LMSLGLSDPT PPSGPSLDGT GWNSFQSSDA TEPPAPALAQ APSMESRPPA QTSLPASSGL 420
421 DDLDLLGKTL LQQSLPPESQ QVRWEKQQPT PRLTLRDLQN KSSSCSSPSS SATSLLHTVS 480
481 PEPPRPPQQP VPTELSLASI TVPLESIKPS NILPVTVYDQ HGFRILFHFA RDPLPGRSDV 540
541 LVVVVSMLST APQPIRNIVF QSAVPKVMKV KLQPPSGTEL PAFNPIVHPS AITQVLLLAN 600
601 PQKEKVRLRY KLTFTMGDQT YNEMGDVDQF PPPETWGSL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
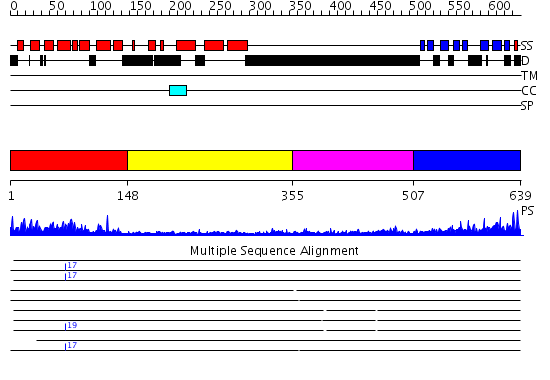
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | 41.522879 | ADP-ribosylation factor binding protein Gga1 | |
| 2 | View Details | [148..354] | 3.81 | ADP-ribosylation factor binding protein Gga1 | |
| 3 | View Details | [355..506] | 1.336996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [507..639] | 26.30103 | ADP-ribosylation factor binding protein Gga1 domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 |
|
||||||||||||
| 2 |
|
||||||||||||
| 3 | No functions predicted. | ||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)