













| General Information: |
|
| Name(s) found: |
OSBPL6
[HGNC (HUGO)]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 934 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSDEKGISP AHKTSTPTHR SASSSTSSQR DSRQSIHILE RTASSSTEPS VSRQLLEPEP 60
61 VPLSKEADSW EIIEGLKIGQ TNVQKPDKHE GFMLKKRKWP LKGWHKRFFV LDNGMLKYSK 120
121 APLDIQKGKV HGSIDVGLSV MSIKKKARRI DLDTEEHIYH LKVKSQDWFD AWVSKLRHHR 180
181 LYRQNEIVRS PRDASFHIFP STSTAESSPA ANVSVMDGKM QPNSFPWQSP LPCSNSLPAT 240
241 CTTGQSKVAA WLQDSEEMDR CAEDLAHCQS NLVELSKLLQ NLEILQRTQS APNFTDMQAN 300
301 CVDISKKDKR VTRRWRTKSV SKDTKIQLQV PFSATMSPVR LHSSNPNLCA DIEFQTPPSH 360
361 LTDPLESSTD YTKLQEEFCL IAQKVHSLLK SAFNSIAIEK EKLKQMVSEQ DHSKGHSTQM 420
421 ARLRQSLSQA LNQNAELRSR LNRIHSESII CDQVVSVNII PSPDEAGEQI HVSLPLSQQV 480
481 ANESRLSMSE SVSEFFDAQE VLLSASSSEN EASDDESYIS DVSDNISEDN TSVADNISRQ 540
541 ILNGELTGGA FRNGRRACLP APCPDTSNIN LWNILRNNIG KDLSKVSMPV ELNEPLNTLQ 600
601 HLCEEMEYSE LLDKASETDD PYERMVLVAA FAVSGYCSTY FRAGSKPFNP VLGETYECIR 660
661 EDKGFRFFSE QVSHHPPISA CHCESKNFVF WQDIRWKNKF WGKSMEILPV GTLNVMLPKY 720
721 GDYYVWNKVT TCIHNILSGR RWIEHYGEVT IRNTKSSVCI CKLTFVKVNY WNSNMNEVQG 780
781 VVIDQEGKAV YRLFGKWHEG LYCGVAPSAK CIWRPGSMPT NYELYYGFTR FAIELNELDP 840
841 VLKDLLPPTD ARFRPDQRFL EEGNLEAAAS EKQRVEELQR SRRRYMEENN LEHIPKFFKK 900
901 VIDANQREAW VSNDTYWELR KDPGFSKVDS PVLW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
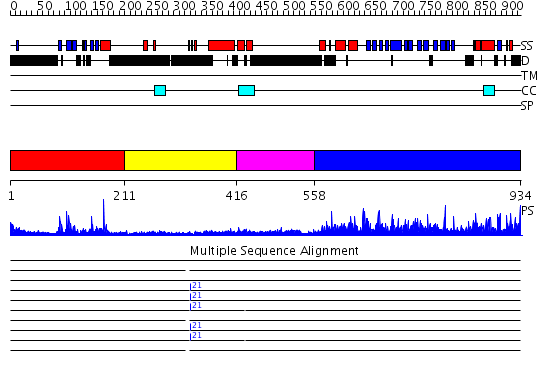
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..210] | 16.39794 | Crystal Structure of a N-terminal Fragment of SKAP-Hom Containing Both the Helical Dimerization Domain and the PH Domain | |
| 2 | View Details | [211..415] | 1.599982 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [416..557] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [558..934] | 80.69897 | Structure of yeast oxysterol binding protein Osh4 in complex with 7-hydroxycholesterol |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)