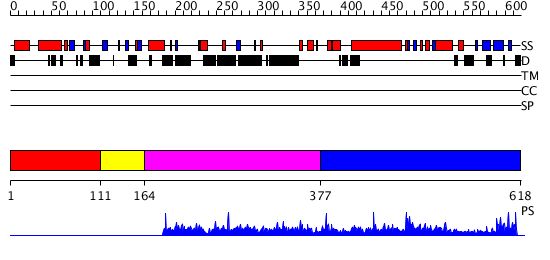
Description(s) found:
SHOW ONLY BEST
|
gi|728707|emb|CAA59391.1| orf 9 [Saccharomyces cerevisiae]
[NCBI NR]
gi|6322358|ref|NP_012432.1| Putative zinc cluster protein of unknown function; proposed to be involved in the regulation of energy metabolism, based on patterns of expression and sequence analysis; Gsm1p [Saccharomyces cerevisiae]
[NCBI NR]
gi|1176484|sp|P42950.1|YJK3_YEAST RecName: Full=Uncharacterized transcriptional regulatory protein YJL103C
[NCBI NR]
pir||S53384 hypothetical protein YJL103c - yeast (Saccharomyces cerevisiae)�
[NCBI NR]
gi|1009448|emb|CAA89398.1| unnamed protein product [Saccharomyces cerevisiae]
[NCBI NR]
Uncharacterized transcriptional regulatory protein YJL103C OS=Saccharomyces cerevisiae GN=YJL103C PE=1 SV=1
[Swiss-Prot]
Glucose starvation modulator protein 1 OS=Saccharomyces cerevisiae GN=GSM1 PE=1 SV=1
[Swiss-Prot]
Similarity to YBR239c
[mips]
Putative zinc cluster protein of unknown function; proposed to be involved in the regulation of energy metabolism, based on patterns of expression and sequence analysis
[SGD]
YJL103C, Chr X from 228722-230578, reverse complement
[yeast_orfs2]
Chr X from 228722-230578, reverse complement
[yeastorf3.fasta]
Chr X from 230578-228722, reverse complement
[yeastorf5.fasta]
(P42950) Putative 70.4 kDa transcriptional regulatory protein in IME2-MEF2 intergenic region
[4932.FASTACCERV.txt]
YJL103C SGDID:S0003639, Chr X from 230798-228942, reverse complement, Uncharacterized ORF
[yeastflysequest.fasta]
Yjl103cp [Saccharomyces cerevisiae]�gi|1009448|emb|CAA89398.1| unnamed protein product [Saccharomyces cerevisiae]�gi|728707|emb|CAA59391.1| orf 9 [Saccharomyces cerevisiae]�gi|1077827|pir||S53384 hypothetical protein YJL103c - yeast (Saccharomyces cerevis
[nrdb_11152004_Chlamydomonas]
YJL103C SGDID:S000003639, Chr X from 230798-228942, reverse complement, Uncharacterized ORF, "Hypothetical protein"
[SGD_S-cerevisiae_na_12-16-2005_con_reversed.fasta]
YJL103C SGDID:S000003639, Chr X from 230877-229021, reverse complement, Uncharacterized ORF, "Putative zinc cluster protein of unknown function; proposed to be involved in the regulation of energy metabolism, based on patterns of expression and sequence a
[yeast-contam.fasta]
GSM1 SGDID:S000003639, Chr X from 230876-229020, reverse complement, Uncharacterized ORF, "Putative zinc cluster protein of unknown function; proposed to be involved in the regulation of energy metabolism, based on patterns of expression and sequence anal
[yeast-200209-contam.fasta]
Putative zinc cluster protein of unknown function; proposed to be involved in the regulation of energy metabolism, based on patterns of expression and sequence analysis; Gsm1p [Saccharomyces cerevisiae]
[nr-271106-contam.fasta]
unnamed protein product [Saccharomyces cerevisiae]
[nr-271106-contam.fasta]
Probable transcriptional regulatory protein YJL103C
[nr-271106-contam.fasta]
orf 9 [Saccharomyces cerevisiae]
[nr-271106-contam.fasta]
GSM1 SGDID:S000003639, Chr X from 230876-229020, reverse complement, Uncharacterized ORF, "Putative zinc cluster protein of unknown function; proposed to be involved in the regulation of energy metabolism, based on patterns of expression and sequence analysis"
[yeast-bovrabprots.fasta]
YJL103C SGDID:S000003639, Chr X from 230877-229021, reverse complement, Uncharacterized ORF, "Putative zinc cluster protein of unknown function; proposed to be involved in the regulation of energy metabolism, based on patterns of expression and sequence analysis"
[yeast-contam_custom.fasta]
[chemostatdb.fasta]
Glucose starvation modulator protein 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) GN=GSM1 PE=1 SV=1
[UniProt_S_cerevisiae_20120516_05-16-2012_reversed.fasta]
GSM1 SGDID:S000003639, Chr X from 230881-229025, Genome Release 64-1-1, reverse complement, Verified ORF, "Putative zinc cluster protein of unknown function; proposed to be involved in the regulation of energy metabolism, based on patterns of expression and sequence analysis"
[yeast-030211-contam.fasta]
GSM1 SGDID:S000003639, Chr X from 230881-229025, Genome Release 64-2-1, reverse complement, Verified ORF, "Putative zinc cluster protein of unknown function; proposed to be involved in the regulation of energy metabolism, based on patterns of expression and sequence analysis"
[20141208_orf_trans.fasta]
YJL103C SGDID:S000003639, Chr X from 230881-229025, Genome Release 64-1-1, reverse complement, Verified ORF, "Putative zinc cluster protein of unknown function; proposed to be involved in the regulation of energy metabolism, based on patterns of expression and sequence analysis"
[2016-01-SGD_Yeast_Contam_Riffle_20140507-Ska-Hec-110-tubulin.fasta]
|