













| General Information: |
|
| Name(s) found: |
STP1 /
YDR463W
[SGD]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 577 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
tRNA splicing, via endonucleolytic cleavage and ligation
[IMP positive regulation of transcription [IDA positive regulation of transcription from RNA polymerase II promoter [IGI |
| Molecular Function: |
transcription factor activity
[IDA specific RNA polymerase II transcription factor activity [IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNCITIDVFF TAFSHPCKRK KKQADSCINC KDSLIKSRFT LQTKGKGKLY RLYHIRSKMP 60
61 STTLLFPQKH IRAIPGKIYA FFRELVSGVI ISKPDLSHHY SCENATKEEG KDAADEEKTT 120
121 TSLFPESNNI DRSLNGGCSV IPCSMDVSDL NTPISITLSP ENRIKSEVNA KSLLGSRPEQ 180
181 DTGAPIKMST GVTSSPLSPS GSTPEHSTKV LNNGEEEFIC HYCDATFRIR GYLTRHIKKH 240
241 AIEKAYHCPF FNSATPPDLR CHNSGGFSRR DTYKTHLKAR HVLYPKGVKP QDRNKSSGHC 300
301 AQCGEYFSTI ENFVENHIES GDCKALPQGY TKKNEKRSGK LRKIKTSNGH SRFISTSQSV 360
361 VEPKVLFNKD AVEAMTIVAN NSSGNDIISK YGNNKLMLNS ENFKVDIPKR KRKYIKKKQQ 420
421 QVSGSTVTTP EVATQNNQEV APDEISSATI FSPFDTHLLE PVPSSSSESS AEVMFHGKQM 480
481 KNFLIDINSF TNQQQQAQDN PSFLPLDIEQ SSYDLSEDAM SYPIISTQSN RDCTQYDNTK 540
541 ISQILQSQLN PEYLSENHMR ETQQYLNFYN DNFGSQF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
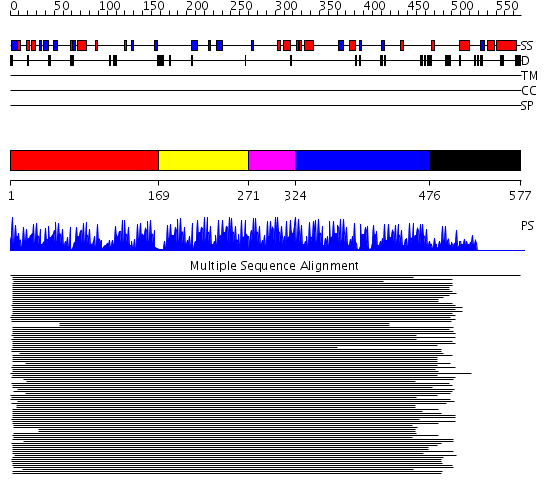
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..168] | 14.019996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [169..270] | 28.0 | Designed zinc finger protein | |
| 3 | View Details | [271..323] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [324..475] | 18.69897 | Ying-yang 1 (yy1, zinc finger domain) | |
| 5 | View Details | [476..577] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)