













| General Information: |
|
| Name(s) found: |
YDR109C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 715 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
kinase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKSRKRQNNM QNETREPAVL SSQETSISRI SPQDPEAKFY VGVDVGTGSA RACVIDQSGN 60
61 MLSLAEKPIK REQLISNFIT QSSREIWNAV CYCVRTVVEE SGVDPERVRG IGFDATCSLV 120
121 VVSATNFEEI AVGPDFTNND QNIILWMDHR AMKETEEINS SGDKCLKYVG GQMSVEMEIP 180
181 KIKWLKNNLE AGIFQDCKFF DLPDYLTFKA TGKENRSFCS AVCKQGFLPV GVEGSDIGWS 240
241 KEFLNSIGLS ELTKNDFERL GGSLREKKNF LTAGECISPL DKKAACQLGL TEHCVVSSGI 300
301 IDAYAGWVGT VAAKPESAVK GLAETENYKK DFNGAIGRLA AVAGTSTCHI LLSKNPIFVH 360
361 GVWGPYRDVL ARGFWAAEGG QSCTGVLLDH LITTHPAFTE LSHMANLAGV SKFEYLNKIL 420
421 ETLVEKRKVR SVISLAKHLF FYGDYHGNRS PIADPNMRAC IIGQSMDNSI EDLAVMYLSA 480
481 CEFISQQTRQ IIEVMLKSGH EINAIFMSGG QCRNSLLMRL LADCTGLPIV IPRYVDAAVV 540
541 FGSALLGAAA SEDFDYTREK RTLKGQKSSQ TKTERFNDSY SSIQKLSMED RNSTNGFVSP 600
601 HNLQLSTPSA PAKINNYSLP ICTQQPLDKT SEESSKDASL TVGQESLGEG RYNGTSFLWK 660
661 VMQELTGNAR IVNPNEKTHP DRILLDTKYQ IFLDMIETQR KYRRMVDKVE GSFSR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
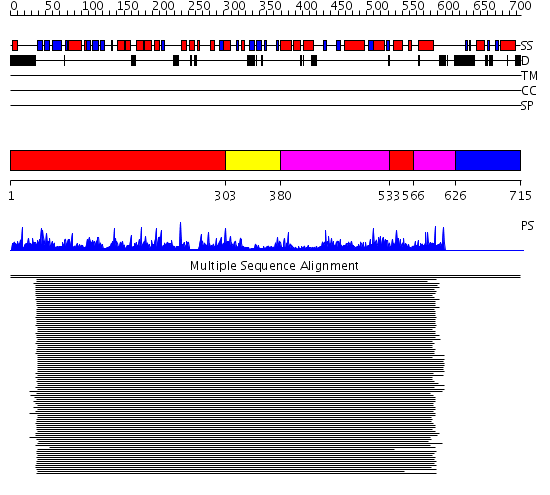
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..302] [533..565] |
190.9897 | Glycerol kinase | |
| 2 | View Details | [303..379] | 190.9897 | Glycerol kinase | |
| 3 | View Details | [380..532] [566..625] |
190.9897 | Glycerol kinase | |
| 4 | View Details | [626..715] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)