













| General Information: |
|
| Name(s) found: |
RDH54 /
YBR073W
[SGD]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 958 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
reciprocal meiotic recombination
[IGI heteroduplex formation [IDA meiotic sister chromatid segregation [IMP double-strand break repair via break-induced replication [TAS |
| Molecular Function: |
DNA-dependent ATPase activity
[IDA DNA topoisomerase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVISVKPRR REKILQEVKN SSVYQTVFDS GTTQMQIPKY ENKPFKPPRR VGSNKYTQLK 60
61 PTATAVTTAP ISKAKVTVNL KRSISAGPTL NLAKKPNNLS SNENTRYFTI MYRKPTTKKH 120
121 KTWSGDGYAT LKASSDKLCF YNEAGKFLGS SMLPSDSDSL FETLFKAGSN EVQLDYELKE 180
181 NAEIRSAKEA LSQNMGNPSP PTTSTTETVP STKNDGGKYQ MPLSQLFSLN TVKRFKSVTK 240
241 QTNEHMTTVP KTSQNSKAKK YYPVFDVNKI DNPIVMNKNA AAEVDVIVDP LLGKFLRPHQ 300
301 REGVKFMYDC LMGLARPTIE NPDIDCTTKS LVLENDSDIS GCLLADDMGL GKTLMSITLI 360
361 WTLIRQTPFA SKVSCSQSGI PLTGLCKKIL VVCPVTLIGN WKREFGKWLN LSRIGVLTLS 420
421 SRNSPDMDKM AVRNFLKVQR TYQVLIIGYE KLLSVSEELE KNKHLIDMLV CDEGHRLKNG 480
481 ASKILNTLKS LDIRRKLLLT GTPIQNDLNE FFTIIDFINP GILGSFASFK RRFIIPITRA 540
541 RDTANRYNEE LLEKGEERSK EMIEITKRFI LRRTNAILEK YLPPKTDIIL FCKPYSQQIL 600
601 AFKDILQGAR LDFGQLTFSS SLGLITLLKK VCNSPGLVGS DPYYKSHIKD TQSQDSYSRS 660
661 LNSGKLKVLM TLLEGIRKGT KEKVVVVSNY TQTLDIIENL MNMAGMSHCR LDGSIPAKQR 720
721 DSIVTSFNRN PAIFGFLLSA KSGGVGLNLV GRSRLILFDN DWNPSVDLQA MSRIHRDGQK 780
781 KPCFIYRLVT TGCIDEKILQ RQLMKNSLSQ KFLGDSEMRN KESSNDDLFN KEDLKDLFSV 840
841 HTDTKSNTHD LICSCDGLGE EIEYPETNQQ QNTVELRKRS TTTWTSALDL QKKMNEAATN 900
901 DDAKKSQYIR QCLVHYKHID PARQDELFDE VITDSFTELK DSITFAFVKP GEICLREQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
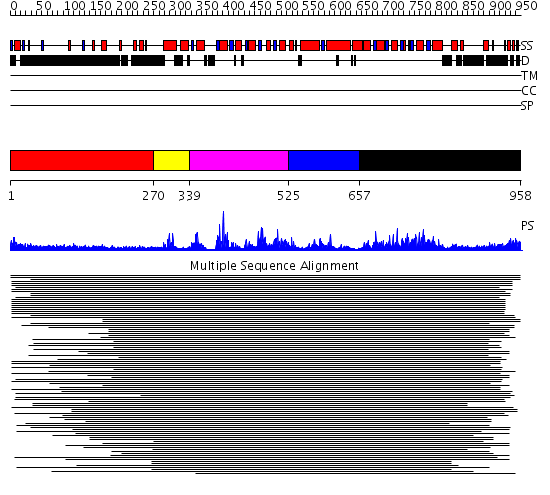
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..269] | 3.282997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [270..338] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [339..524] | 11.96 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 4 | View Details | [525..656] | 19.39794 | Putative DEAD box RNA helicase | |
| 5 | View Details | [657..958] | 5.69897 | Nucleotide excision repair enzyme UvrB |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)