













| General Information: |
|
| Name(s) found: |
SYP1_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 818 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESLTKTEYV DAFLSNYSPN DSMSIFRQRL EQVRLDNDML SQWIRERMDI ERQYSDQLHK 60
61 LAMNMQEKNN SSFAFNYAWK QLEGETLEIS RYHSQIIGQI ASQVYKPLID YYTSSPQTAT 120
121 LRRLAERLST VAEEMASSSV PGLKGLKKKG RDADTKSQND LTASRATWDS DAPLAFEKLQ 180
181 IVDEERLLIL KQVYLTIASL ETDTALKQQE FFSKSMAVYS DLPIEGEIRQ FMNSTSKVMS 240
241 SASANKPSKS SGFHINNGKS KEEKSHGENE SGGKLKNKMS TLFRRKTIMP KKDKKPSHKS 300
301 NGRPNKLTAF FNKNSKASSI SSAEEHPSNI DDSSIERRHY DSNHSSQIRD HPSTNNNASS 360
361 YQNFNETSDE GEDNDATIRA NNVRSSFLEA PLPVQPNVQA ETVTPKISSK ASPFNKPNSV 420
421 HSEASRNTPS SIDRENASHS NPIMMHGNDF GGNFNNMQSR STTTSPTSAV ASPAPTENED 480
481 SNAAIERVAN TLRKNPTISR RTRRAGTMDR YATASSDYME SNLGSLPNLS TLSLQSGPDS 540
541 TATWHPEFPN SSGLSASIVE KYSGELSEDG LLHPSCSGHI FMKYTSDFNT PPPEMGVRVA 600
601 SEFPMSFTHL NDHAVKYGPS ENTLSLIPEL LLSPIKVTDF NLHLDSINGA SCIPLTVVQK 660
661 WKHDESSSSM IAFVKPNPVW RNLGSSLHIE KLVIIVYLGE NVLVKSCQSS PAGEFSRKTS 720
721 KLKLHLSNIN ISSSGFKILA KFAISPSAAI RKPVIEFRIR MVDSSNNPGL TKLFLKPDMF 780
781 DTTSSSGGSQ LESAYEETKV PTSYGIHVRE CSVFADMS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
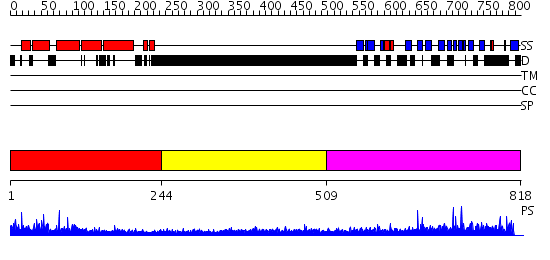
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..243] | 57.69897 | No description for 2v0oA was found. | |
| 2 | View Details | [244..508] | 2.296996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [509..818] | 1.74 | Second domain of Mu2 adaptin subunit (ap50) of ap2 adaptor; Mu2 adaptin (clathrin coat assembly protein AP50) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)