













| General Information: |
|
| Name(s) found: |
gi|56202337
[NCBI NR]
gi|215697038 [NCBI NR] gi|115446539 [NCBI NR] gi|113536580 [NCBI NR] |
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 475 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGSGRPPAA QKKILQSLRP PLPFAASSRS PFAAPNDYHR FPAGGAAAAA ASGSGGIGAG 60
61 GAGGGGDIEE GLVIRTPQKR KAPEESDVAE SSDCMITSPG FAVSPMLTPV SGKAVKTSKS 120
121 KTKNNKAGPQ TPTSNVGSPL NPPTPVGTCR YDSSLGLLTK KFINLLKQAP DGILDLNNAA 180
181 ETLEVQKRRI YDITNVLEGI GLIEKTLKNR IRWKGLDDSG VELDNGLSAL QAEVENLSLK 240
241 EQALDERISD MREKLRGLTE DENNQRWLYV TEDDIKGLPC FQNETLIAIK APHGTTLEVP 300
301 DPDEAGDYLQ RRYRIVLRST MGPIDVYLVS QFDEKFEDLG GGATPSGHAN VPKHQPTEVF 360
361 NTTNAGVGQC SNSVAVDNNI QHSQTIPQDP SASHDFGGMT RIIPSDIDTD ADYWLISEGD 420
421 VSITDMWKTA PDVQWDESLD TDVFLSEDVR TPSSHNQQPS AVGGPQMQVS DMHKP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
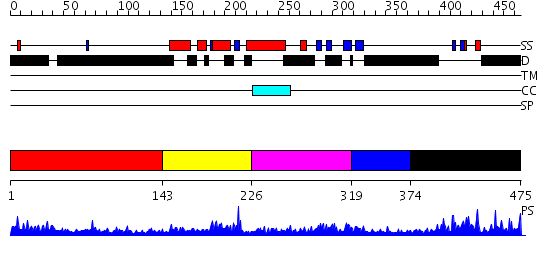
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..142] | 1.11799 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [143..225] | 23.221849 | Cell cycle transcription factor E2F-4 | |
| 3 | View Details | [226..318] | 5.05 | Structure of the Rb C-terminal domain bound to an E2F1-DP1 heterodimer | |
| 4 | View Details | [319..373] | 24.045757 | Structure of the Rb C-terminal domain bound to an E2F1-DP1 heterodimer | |
| 5 | View Details | [374..475] | 2.079996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)