













| General Information: |
|
| Name(s) found: |
gi|62471665
[NCBI NR]
gi|61678346 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1998 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
signal transduction
[IEA]
actin filament organization [ISS] filopodium assembly [IEA] |
| Molecular Function: |
cytoskeletal adaptor activity
[IEA]
actin binding [ISS] SH3 domain binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLSLERDSS ALGSLFQQII NDMKNTSPLW EDFVAKAGKL HTCLRAAIQA IAAYLDAFQK 60
61 IADAATNSRG ASKEIGTALT RVCLRHKAVE TRLKTFTSTI MDCLVQPLQE RIEDWKRTVA 120
121 TIDKDHAKEY KRCRSELKKR SSDTLRLQKK ARKGQTDGLQ SLMDSHMQDV TLRRAELEEV 180
181 EKKSLRAAMV EERLRYCSFV HMLQPVVHEE CEVMSELGHL QEAMQSIALV TKEPSVLPQA 240
241 SEELIHDAKA SINLYPESPG GGSGSQGGGC SNSLGSRKSS VCSISSMNSS GSSNSPGHHH 300
301 YPRSLSQTSN ATNQTANVST WPPHSQDGVD TLPPTADRPH TISTAYEKGH QRPPLTVYTF 360
361 QNPETIHESG SCLNNGTAAP NGQPLSGQAT PATQKSPAAS LSRPPLPVRC SSLERPLSAQ 420
421 SNHRQGSGNN LLQRQCPSPI PAHITKELSA AHHAQQQQQQ QNQQPQTPPT YVNMSELATM 480
481 AALKQTNQQQ KPSTPPLQQQ SSIDSTSSQH SNDSTGSHQL LQQQQQHHQS QQNHHSATAT 540
541 RSHSISSTAS SLHSHPSIDS TVACGSLVGQ HNHSTSTNTN TTSPSSGSST PQNHYSPLLT 600
601 NSPTSTAAGT PSGSSLGPGS GLGFVYQVSS PTPPSSEVLK ITEQAAAGQD QGPANSVADE 660
661 TDERSRASVL QKASMFEKAA AAAAVSPPAP MQIASGSPAS GGGTRRSEAE QQEMDKSFED 720
721 SIQALNNLIG ELDSFQREID EGKVKPPSNI ISGSTTSSNN NNTTTSSISS SDNNNLPATS 780
781 NIEPCAISNQ TNSSGCGTDI SDTTSDELAG DDMDVRRRDR DRDRDLLGAS DSELSRCYVS 840
841 ETSSLTGGLT AGGYENPTFA HFAANANRED AVSLASDSVC LGQPRHAYVD TCSDSGSAVV 900
901 VIYDHQIPNT PDIEFVKQNS EIVVLRTKDP QPHALQLHEM RELQQLPANL AGSPDSSPDS 960
961 AGGQAPPTAT VAPAKQRLSS FRATSEQQLQ LLGRGSPQRG KTPSEQAVQS RPQDQHFPQT1020
1021 QQQDIDGSSP PVELARRQLP PKPTSLSIFN GPVPTAGDRP VVPRKSDFKA DLDAKIRRQK1080
1081 QKVKQQLQQQ QQQQQQEQQE TQQQQQAPQE QQHSPQSPQT RNCNVTNQQA ANITASASAF1140
1141 ATATASTDPY PNPNHRMPNQ NQTATSNHTQ CKTPTMALSP SSPRGHLPLS SSSLSSLPLP1200
1201 ATTSSPSNAR PSMLPASDRP PAHPYVCSNA PANPHHANSI SNANAHLKPC ITPRPASLSG1260
1261 GAAGGGSTRI GRRSSINQSK PPPPVRRSSS VTPSPNASVG LQQQPQHATL SQQNHQLSSS1320
1321 SEHLPPPPPF MLDAMPQIPS SALKVSETVR ALAAMRHQPA SPVSLRRMQQ QQQQQLQQQQ1380
1381 QQQVQQQQPL LQSAHNSPLK EDLTVYYDSY LDLHAYAQAL ASGQQPGGQQ MANQQRFTLQ1440
1441 QQHQHLHQPQ QPPVYQVDAT FRTSSPAAGG GGGGGIYAQP KLVNSMSSFR TSSPSPNGHA1500
1501 HPLPPTQPKA NPNLIAQLNA RLSGKQQQHQ QQQHIEGIYG NQQAPGGESI YMRSGLSMSQ1560
1561 PQQQQHFDGK SEQIQLQHQQ QHRIYASFGT SSSAMSSAHA ANSSTKPSIL TPTTSFNALP1620
1621 HFPLSSSTSS LLSKVSSFSN SSSASPPTTA ATSGSASSHY QPPQPPNAAV ANSKDMAIYS1680
1681 SSFTKNPAAA QSPNMRQAHS HQHHQPPQQQ HYTCPPPLED PPPPPIYAAG ASATMPKKMA1740
1741 RPPTGQNATH SSAYAAASST ATLPKNMMQQ QQRLQHQQQY QQPAGMGIGN GNGHLGQRPQ1800
1801 LPLPQQKLRA AQQQHLAEQQ HQQQQEQQQH HQQHQQHQQR QPPIPSRHSS VQQKIFVSTN1860
1861 PFIQTTAVKF HSPSASPTCG SPVTGSLASI YATTSRGGHH HQQQQAQQQQ HYYRDAAGGN1920
1921 SNGGAAYYNH NAHAHSQAHH PNWQYSCQDQ GRIPRESQRE AGEAGNVWTS ICRAKSHQQQ1980
1981 GPDVSKSAKT IATQCAIP |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
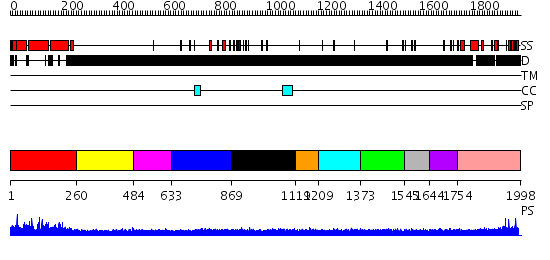
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..259] | 123.0 | No description for 2d1lA was found. | |
| 2 | View Details | [260..483] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [484..632] | 1.108984 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [633..868] | 2.121994 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [869..1118] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1119..1208] | 2.127986 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1209..1372] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1373..1544] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1545..1643] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1644..1753] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1754..1998] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)