













| General Information: |
|
| Name(s) found: |
CG11727-PA /
FBpp0073392
[FlyBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 807 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: |
regulation of Rab GTPase activity
[IEA]
|
| Molecular Function: |
Rab GTPase activator activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTLTTTTTAS SAESQAKMDV KGGALPGEEN LPTSEMDLLA KLEAANKLIE SDAKSLNSLH 60
61 STHSRKNSDT SQISLTSSGN SVAEEDIWTT WATILNDWEG ALKRKNPCVS ELVRRGIPHH 120
121 FRAIVWQQLS GASDGDKKQY AEYIKATSAC EKVIRRDIAR TYPEVEFFKE KDGPGQEALF 180
181 NVIKAYSLHD REVGYCQGSG FIVGLLLMQM PEEEAFAVLV QIMQQHRMRH MFKPSMSELG 240
241 LCMYQLENLV QEQIPDMHIH FQQQGFQTTM YASSWFLTLY TTTLNVNLSC RIMDVFLSEG 300
301 MEFIFKVALA LLLTGKDTLL CLDMEAMLKF FQKELPGRVE ADVEGFFNLA YSIKLNTKRM 360
361 KKMEKEYQDL KKKEQEEMAE LRRLRRENCL LKQRNELLEA ESAELADRLV RGQVSRAEEE 420
421 ETSYAIQTEL MQLRRSYLEV SHQLENANEE VRGLSLRLQE NNVSIDSNNS RQSSIDELCM 480
481 KEEALKQRDE MVSCLLEELV KVRQGLAESE DQIRNLKAKV EELEEDKKTL RETTPDNSVA 540
541 HLQDELIASK LREAEASLSL KDLKQRVQEL SSQWQRQLAE NQRSESERTT NAVDSTPKKL 600
601 LTNFFDSSKS SEHTQKLEEE LMTTRIREME TLTELKELRL KVMELETQVQ VSTNQLRRQD 660
661 EEHKKLKEEL EMAVTREKDM SNKAREQQHR YSDLESRMKD ELMNVKIKFT EQSQTVAELK 720
721 QEISRLETKN SEMLAEGELR ANLDDSDKVR DLQDRLADMK AELTALKSRG KFPGAKLRSS 780
781 SIQSIESTEI DFNDLNMVRR GSTELST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
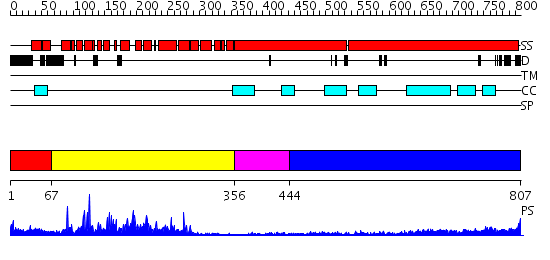
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..66] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [67..355] | 50.30103 | No description for 2qq8A was found. | |
| 3 | View Details | [356..443] | 1.16 | No description for 2fxmA was found. | |
| 4 | View Details | [444..807] | 20.69897 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)