













| General Information: |
|
| Name(s) found: |
gi|61356797
[NCBI NR]
gi|61356792 [NCBI NR] gi|168277782 [NCBI NR] gi|157928502 [NCBI NR] gi|123993239 [NCBI NR] |
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | synthetic construct |
| Length: | 543 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEQVNELKEK GNKALSVGNI DDALQCYSEA IKLDPHNHVL YSNRSAAYAK KGDYQKAYED 60
61 GCKTVDLKPD WGKGYSRKAA ALEFLNRFEE AKRTYEEGLK HEANNPQLKE GLQNMEARLA 120
121 ERKFMNPFNM PNLYQKLESD PRTRTLLSDP TYRELIEQLR NKPSDLGTKL QDPRIMTTLS 180
181 VLLGVDLGSM DEEEEIATPP PPPPPKKETK PEPMEEDLPE NKKQALKEKE LGNDAYKKKD 240
241 FDTALKHYDK AKELDPTNMT YITNQAAVYF EKGDYNKCRE LCEKAIEVGR ENREDYRQIA 300
301 KAYARIGNSY FKEEKYKDAI HFYNKSLAEH RTPDVLKKCQ QAEKILKEQE RLAYINPDLA 360
361 LEEKNKGNEC FQKGDYPQAM KHYTEAIKRN PKDAKLYSNR AACYTKLLEF QLALKDCEEC 420
421 IQLEPTFIKG YTRKAAALEA MKDYTKAMDV YQKALDLDSS CKEAADGYQR CMMAQYNRHD 480
481 SPEDVKRRAM ADPEVQQIMS DPAMRLILEQ MQKDPQALSE HLKNPVIAQK IQKLMDVGLI 540
541 AIR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
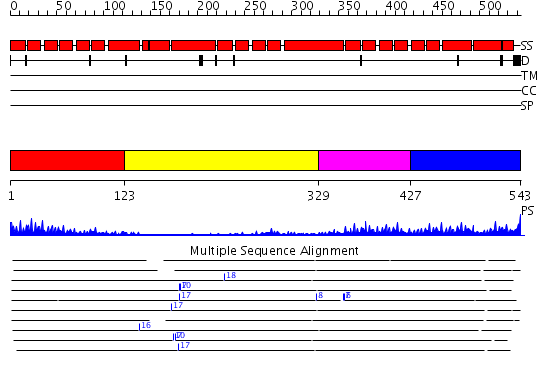
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | 12.09691 | Hop | |
| 2 | View Details | [123..328] | 5.43 | No description for 2q7fA was found. | |
| 3 | View Details | [329..426] | 5.522879 | Neutrophil cytosolic factor 2 (NCF-2, p67-phox) | |
| 4 | View Details | [427..543] | 4.0 | Crystal Structure of the Hypothetical protein VPA1032 from Vibrio parahaemolyticus, Northeast Structural Genomics Target VpR44 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)