













| General Information: |
|
| Name(s) found: |
gi|83584790
[NCBI NR]
gi|194439800 [NCBI NR] gi|194421248 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli 101-1 |
| Length: | 581 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGRIPRVFI NDLLARTDIV DLIDARVKLK KQGKNFHACC PFHNEKTPSF TVNGEKQFYH 60
61 CFGCGAHGNA IDFLMNYDKL EFVETVEELA AMHNLEVPFE AGSGPSQIER HQRQTLYQLM 120
121 DGLNTFYQQS LQQPVATSAR QYLEKRGLSH EVIARFAIGF APPGWDNVLK RFGGNPENRQ 180
181 SLIDAGMLVT NDQGRSYDRF RERVMFPIRD KRGRVIGFGG RVLGNDTPKY LNSPETDIFH 240
241 KGRQLYGLYE AQQDNAEPNR LLVVEGYMDV VALAQYGINY AVASLGTSTT ADHIQLLFRA 300
301 TNNVICCYDG DRAGRDAAWR ALETALPYMT DGRQLRFMFL PDGEDPDTLV RKEGKEAFEA 360
361 RMEQAMPLSA FLFNSLMPQV DLSTPDGRAR LSTLALPLIS QVPGETLRIY LRQELGNKLG 420
421 ILDDSQLERL MPKAAESGVS RPVPQLKRTT MRILIGLLVQ NPELATLVPP LENLDENKLP 480
481 GLGLFRELVN TCLSQPGLTT GQLLEHYRGT NNAATLEKLS MWDDIADKNI AEQTFTDSLN 540
541 HMFDSLLELR QEELIARERT HGLSNEERLE LWTLNQELAK K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
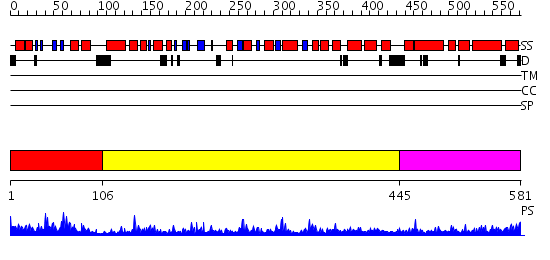
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..105] | 35.154902 | Zinc-binding domain of DNA primase | |
| 2 | View Details | [106..444] | 83.0 | DNA primase DnaG catalytic core | |
| 3 | View Details | [445..581] | 27.0 | Crystal Structure of the E.coli DnaG C-terminal domain (residues 434 to 581) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)