













| General Information: |
|
| Name(s) found: |
YD14_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 638 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLVELDYLS ALEDLTFNSK PIIHTLTYIA QENEPYAISI VNAIEKHIQK CPPNCKLPAL 60
61 YLLDSISKNL GAPYTYFFGL HLFSTFMSAY TVVEPRLRLK LDQLLATWKQ RPPNSSSLEP 120
121 VFSPIVTAKI ENALLKYKST ILRHQSPLLA NTSISSFSAP IDANINSYSS FSDPASSYKP 180
181 SLPSVPFGFQ HISGTSPSPG FITLDSLLSD VNRMIVTEQA RFIKNPYDNM AKKRFEILLQ 240
241 LKNVLSSSAL PYDQLLAIKN QLAQLEKPAS PSTSSVATSA PSVPSALSSI SSTPFMKPSI 300
301 PSTIPTIPSA YSASVSSQPP LTHSYVHPGP QSHKYSLSSG PPASLYNANA LTPEESSSID 360
361 SLFANLQAAG LVPPSAGGKS QGPQASCTEA VSLTADIDLS KSSLATPRPK LSSLLYENYS 420
421 NQCANCGRRY GNDPESRKEL DKHSDWHFRI NKRIRESSLH GINRCWFVME EEWVNSKEEE 480
481 DLITETAQEI EEQRQKQMES VRSQYVLTPL DPIAASEPCP ICQEKFQSVW HEEAEVWVFM 540
541 NAVEEEGRIF HATCLQEVRP SENKHSNTNT STQNLAEAAV SSNIKNATGD ASKDSQPDVQ 600
601 NLLQGIDIQS ILQALGKRKE RDDSMDSMSS KVIKQESK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
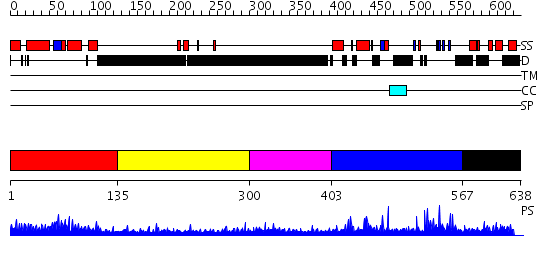
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | 43.39794 | The RNA polymerase II CTD in mRNA processing: beta-turn recognition and beta-spiral model | |
| 2 | View Details | [135..299] | 3.045757 | Sec24 | |
| 3 | View Details | [300..402] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [403..566] | 2.1 | No description for 2i13A was found. | |
| 5 | View Details | [567..638] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)