













| General Information: |
|
| Name(s) found: |
dcap-2 /
CE39374
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 786 amino acids |
Gene Ontology: |
|
| Cellular Component: |
P granule
[IDA cytoplasmic mRNA processing body [IDA |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP]
|
| Molecular Function: |
hydrolase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEISTENWCK KPKNRSIFSK NISFQKQNKS TEEPPSSVQK LLASLQQAQN KSDLSEQPST 60
61 SKPKKNEKRK KAVAQAPASA PAPGPEEKKK QPKRASVGAR MQQQAENARI SQTKRPRQVS 120
121 TSKGSSRNTT APEQQNYQQQ QQQYKGPRIP TDILDELEFR FISNMVECEI NDNIRVCFHL 180
181 ELAHWYYIDH MVEDDKISGC PNVGSRDFNF QMCQHCRVLR KYAHRADEVL AKFREYKSTV 240
241 PTYGAILVDP EMDHVVLVQS YFAKGKNWGF PKGKINQAEP PRDAAIRETF EETGFDFGIY 300
301 SEKEKKFQRF INDGMVRLYL VKNVPKDFNF QPQTRKEIRK IEWFKIDDLP TDKTDELPAY 360
361 LQGNKFYMVM PFVKDIQIYV QKEKEKLRRR KAEAVQSTPS SSIFSQLFPA QPPPPVPEDA 420
421 TPTRPMYKRL TSEELFSAFK NPPAGEVARP TLPDMSPAVN GLDTLAVLGI CTPLKPGASL 480
481 NEFSGAPQNC PMISEEAGSP ADPSAEIGFA MPMDLKQPVV TSDHPWQHHK ISDSSAPPQT 540
541 LESHQGWLDT QLVNTIMHSP NHPLPPTSNS PATPTAVLGH LIGKPIQPQA ILPQAATPTA 600
601 LGSAEKPKSS RISLSDNSAF KAISSTQKQS IPKATAAPPS TEKTRSASLS GSSQVVGKPA 660
661 RNLFNSVVSP VSSGIQSIQG DGGAWEDVWF REQLAATTTA GTSISSLAAS NQELAMINRE 720
721 ETPIEDPYFK QQAYQKAQKA QSLIPACSQW TNSIKLDIDY VVGPLSFWMQ QFSTKSPVSG 780
781 TGPQLP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
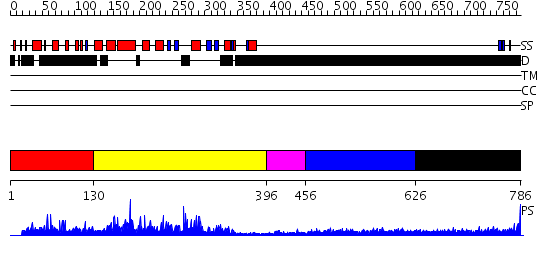
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..129] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [130..395] | 89.0 | Crystal structure of S.pombe mRNA decapping enzyme Dcp2p | |
| 3 | View Details | [396..455] | 11.39794 | RBP1 | |
| 4 | View Details | [456..625] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [626..786] | 3.270995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)