













| General Information: |
|
| Name(s) found: |
PRR73_ORYSA
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Oryza sativa |
| Length: | 767 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSACEAGTD EPSRDDVKGT GNGILENGHS HKPEEEEWRN GMGEDLPNGH STPPEPQQTD 60
61 EQKEHQVQIV RWERFLPVKT LRVLLVENDD STRQVVSALL RKCCYEVIPA ENGLHAWQCL 120
121 EDLQNHIDLV LTEVVMPRLS GIGLLSKITS HKICKDIPVI MMSSNDSMGT VFKCLSKGAV 180
181 DFLVKPIRKN ELKNLWQHVW RRCHSSSGSG SESGIRTQKC TKPKVDDEYE NNSGSNNDNE 240
241 DDDDNDEDDD DLSVGHNARD GSDNGSGTQS SWTKRAVEID SPQQMSPDQP SDLPDSTCAQ 300
301 VIHPTSEICS NRWLPTANKR SGKKHKENND DSMGKYLEIG APRNSSMEYQ SSPREMSVNP 360
361 TEKQHETLMP QSKTTRETDS RNTQNEPTTQ TVDLISSIAR STDDKQVVRI NNAPDCSSKV 420
421 PDGNDKNRDS LIDMTSEELG LKRLKTTGSA TEIHDERNIL KRSDLSAFTR YHTTVASNQG 480
481 GAGFGGSCSP QDNSSEALKT DSNCKVKSNS DAAEIKQGSN GSSNNNDMGS STKNAITKPS 540
541 SNRGKVISPS AVKATQHTSA FHPVQRQTSP ANVVGKDKVD EGIANGVNVG HPVDVQNSFM 600
601 QHHHHVHYYV HVMTQQQQQP SIERGSSDAQ CGSSNVFDPP IEGHAANYSV NGSFSGGHNG 660
661 NNGQRGPSTA PNVGRPNMET VNGIVDENGA GGGNGSGSGS GNDLYQNGVC YREAALNKFR 720
721 QKRKVRNFGK KVRYQSRKRL AEQRPRIRGQ FVRQSGQEDQ AGQDEDR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
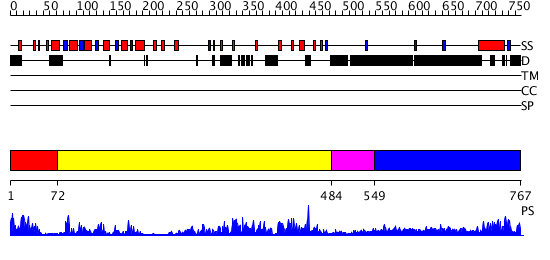
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] | 15.468994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [72..483] | 87.0 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 3 | View Details | [484..548] | 72.69897 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding | |
| 4 | View Details | [549..767] | 11.036978 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)