













| General Information: |
|
| Name(s) found: |
NCKP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1425 amino acids |
Gene Ontology: |
|
| Cellular Component: |
SCAR complex
[TAS]
|
| Biological Process: |
multidimensional cell growth
[IMP]
actin nucleation [TAS] actin filament organization [IMP] trichome morphogenesis [IMP] |
| Molecular Function: |
transcription activator activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANSRQYYPS QDESMSPTSV RSREWEGPSR WTEYLGPEMA ASVSSTRSSK QIDGHVGGST 60
61 KALNIQWVVQ MIEVADGLMA KMYRLNQILE YPDPVGHVFS EAFWKAGVFP NHPRICTLLS 120
121 KKFPEHFSKL QLERIDKFSL DSLHDGAELH LQSLEPWIQL LLDLMAFREQ ALRLILDLSS 180
181 TVITLLPHQN SLILHAFMDL FCAFVRVNLF AEKIPRKMLL QVYNLLHALS RNDRDCDFYH 240
241 RLVQFIDSYD PPLKGLQEDL NFVSPRIGEV LEAVGPSIFL SADTRKLRNE GFLSPYHPRF 300
301 PDILTNSAHP MRAQDLANVT SYREWVLLGY LVCPDELLRV TSIDIALVVL KENLVVTLFR 360
361 DEVSLYQMVC EKEFGIGISF ASADSINLTM QYILLHEDYQ LYVLPRVLES KKMAKSGRTK 420
421 QKEADLEYSV AKQVEKMISE VHEQALQLCD TIHRERRILL KQEIGRMVLF FTDQPSLLAP 480
481 NIQMVFSALA LAQSEVLWYF QHAGIASSRS KAARVIPVDI DPNDPTIGFL LDGMDRLCCL 540
541 VRKYISAARG YALSYLSSSA GRIRYLMGTP GIVALDLDPT LKGLFQRIVQ HLESIPKAQG 600
601 ENVSAITCDL SDFRKDWLSI LMIVTSSRSS INIRHLEKAT VSTGKEGLLS EGNAAYNWSR 660
661 CVDELESQLS KHGSLKKLYF YHQHLTTVFR NTMFGPEGRP QHCCAWLSVA SSFPECASLI 720
721 IPEEVTKFGR DAVLYVESLI ESIMGGLEGL INILDSEGGF GALESQLLPE QAAAYLNNAS 780
781 RISAPSVKSP RVVGGFTLPG HESYPENNKS IKMLEAAIQR LTNLCSILND MEPICVINHV 840
841 FVLREYMREC ILGNFKRRFL TALQTDNDLQ RPSVLESLIR RHMGIVHLAE QHVSMDLTQG 900
901 IREILLTEAF SGPVSSLHTF EKPAEQQQTT GSAVEVVCNW YMDNIIKDVS GAGILFAPRH 960
961 KYFKSTRPVG GYFAESVTDL KELQAFVRIF GGYGVDRLDR MMKVHTAALV NCIETSLRSN1020
1021 RELIEAAAAS MHSGDRVERD ASVRQIVDLD TVIGFCIEAG QALAFDDLLA EASGAVLEDN1080
1081 ASLIHSMISG IVEHIPEEIP EKKEIRRIKG VANGVGVAGD HDSEWVRLIL EEVGGANDNS1140
1141 WSLLPYFFAS FMTSNAWNTT GFNIETGGFS NNIHCLARCI SAVIAGSEYV RLQREYQQQH1200
1201 QSLSNGHHSS ENLDSEFPPR VTAEASIKSS MLLFVKFAAS IVLDSWSEAN RSHLVAKLIF1260
1261 LDQLCEISPY LPRSSLESHV PYTILRSIYT QYYSNTPSTP LSTASPYHSP SVSLIHASPS1320
1321 MKNSTTPQRG SGSGSSSTAA PDSGYFKGSS SSLYGQEHYT ESETGNSRNN ENNNNNKQRG1380
1381 SSRRSGPLDY SSSHKGGSGS NSTGPSPLPR FAVSRSGPIS YKQHN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
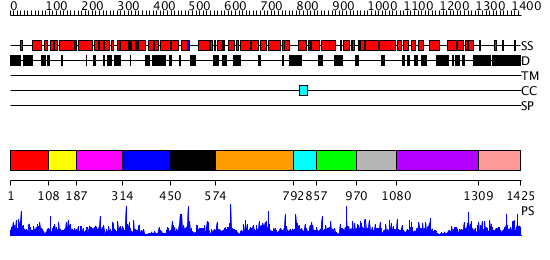
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..107] | 1.015962 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [108..186] | 1.016994 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [187..313] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [314..449] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [450..573] | 1.022956 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [574..791] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [792..856] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [857..969] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [970..1079] | 1.031969 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1080..1308] | 1.030962 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1309..1425] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)