













| General Information: |
|
| Name(s) found: |
CAPP2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 963 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
apoplast [IDA] cytosol [IDA] plasma membrane [IDA] |
| Biological Process: |
tricarboxylic acid cycle
[ISS]
|
| Molecular Function: |
catalytic activity
[IEA]
phosphoenolpyruvate carboxylase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAARNLEKMA SIDAQLRLLA PGKVSEDDKL IEYDALLLDR FLDILQDLHG EDVREFVQEC 60
61 YEVAADYDGN RNTEKLEELG NMLTSLDPGD SIVVTKSFSN MLSLANLAEE VQIAYRRRIK 120
121 KLKKGDFADE ASATTESDIE ETLKRLLQLN KTPEEVFDAL KNQTVDLVLT AHPTQSVRRS 180
181 LLQKFGRIRD CLTQLYAKDI TPDDKQELDE ALQREIQAAF RTDEIRRTPP TPQDEMRAGM 240
241 SYFHETIWKG VPKFLRRVDT ALKNIGINER VPYNAPLIQF SSWMGGDRDG NPRVTPEVTR 300
301 DVCLLARMMA ANLYFSQIED LMFEMSMWRC NEELRVRAER QRCAKRDAKH YIEFWKQIPA 360
361 NEPYRAILGD VRDKLYNTRE RARQLLSSGV SDVPEDAVFT SVDQFLEPLE LCYRSLCDCG 420
421 DRPIADGSLL DFLRQVSTFG LALVKLDIRQ ESERHSDVLD AITTHLGIGS YKEWSEDKRQ 480
481 EWLLSELSGK RPLFGPDLPK TEEVADVLDT FKVISELPSD SFGAYIISMA TAPSDVLAVE 540
541 LLQRECGITD PLRVVPLFEK LADLESAPAA VARLFSIEWY RNRINGKQEV MIGYSDSGKD 600
601 AGRLSAAWQL YKTQEELVKV AKEYGVKLTM FHGRGGTVGR GGGPTHLAIL SQPPDTIHGQ 660
661 LRVTVQGEVI EQSFGEEHLC FRTLQRFTAA TLEHGMHPPV SPKPEWRVLM DEMAIIATEE 720
721 YRSVVFKEPR FVEYFRLATP ELEYGRMNIG SRPSKRKPSG GIESLRAIPW IFAWTQTRFH 780
781 LPVWLGFGGA FKRVIQKDSK NLNMLKEMYN QWPFFRVTID LVEMVFAKGD PGIAALYDRL 840
841 LVSEELQPFG EQLRVNYQET RRLLLQVAGH KDILEGDPYL RQRLQLRDPY ITTLNVCQAY 900
901 TLKQIRDPSF HVKVRPHLSK DYMESSPAAE LVKLNPKSEY APGLEDTVIL TMKGIAAGMQ 960
961 NTG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
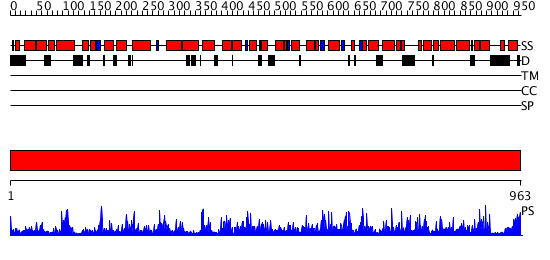
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..963] | 1000.0 | Phosphoenolpyruvate carboxylase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)