













| General Information: |
|
| Name(s) found: |
dab-1 /
CE37844
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 492 amino acids |
Gene Ontology: |
|
| Cellular Component: |
AP-1 adaptor complex
[IDA vesicle [IDA AP-2 adaptor complex [IDA Golgi-associated vesicle [IDA |
| Biological Process: |
molting cycle, protein-based cuticle
[IMP vesicle-mediated transport [IEP locomotory behavior [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP oviposition [IMP protein secretion [IMP cell migration [IMP positive regulation of locomotion [IMP |
| Molecular Function: |
clathrin heavy chain binding
[IDA protein domain specific binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQKSDISVE TANATSGKPN PPSPKSRLAM LKRTKKASNA SSDPFRFQNN GISYKGKLIG 60
61 EQDVDKARGD AMCAEAMRTA KSIIKAAGAH KTRITLQINI DGIKVLDEKS GAVLHNFPVS 120
121 RISFIARDSS DARAFGLVYG EPGGKYKFYG IKTAQAADQA VLAIRDMFQV VFEMKKKQIE 180
181 QVKQQQIQDG GAEISSKKEG GVAVADLLDL ESELQQIERG VQQLSTVPTN CDAFGASPFG 240
241 DPFVDSFNST ATSNGTANMS GTQVPFGGLQ LPQVQQMQMP MVQIPQQSHQ NWPTSGAGSF 300
301 DAWGQQQQQQ QMHHAHSTPA FGTNGFSDTN PFASAFNTQA RPPPLPTVAP TQYRDPFSVH 360
361 SSAIPNSNID WTGTGTTKEN MAPSTNIQQQ QSSLHHASTF ANFGDNKFRA ETWSEKKVTS 420
421 LEEAFTKLVD MDALVGGQGI KETKKNPFEH ILNPPKASLN SMSTTCSAAQ MAATQQHTTS 480
481 SHADPFGDDF FR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
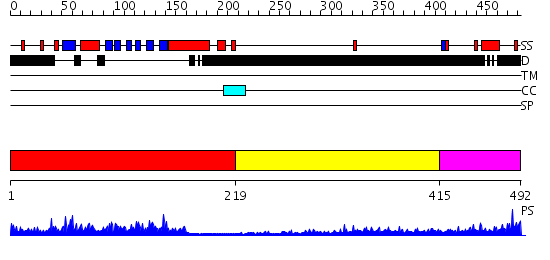
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..218] | 47.154902 | Crystal structure of the phosphotyrosine binding domain (PTB) of mouse Disabled 1 (Dab1) | |
| 2 | View Details | [219..414] | 2.190996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [415..492] | 1.16299 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)