













| General Information: |
|
| Name(s) found: |
CE38201
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 786 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
nematode larval development
[IMP embryonic development ending in birth or egg hatching [IMP positive regulation of growth rate [IMP rRNA processing [IEA growth [IMP reproduction [IMP |
| Molecular Function: |
rRNA (uridine) methyltransferase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKKVKIGKQ RRDKYYKLAK EAGYRSRAAF KLVQLNKRFE FLEKSRATVD LCAAPGGWMQ 60
61 VASQFMPVSS LIVGVDLAPI KPIKNCIALQ GDITTNETRA AIKKELKTWS ADCVLHDGAP 120
121 NVGLNWVHDA FQQNCLTLSA LKLATQILRK GGTFVTKVFR SNDYSCLIRV FEKLFKRVHV 180
181 WKPAASRLES AEIFVVCEVY QKPDKVGAEY LDPKKVFANP DGSEGTKPNP QNLLIGKQKK 240
241 AKAEGYDTDS LAIHSTINAT DFIKSSGYLD ILGTANVITL DDEKWKNHEK TTEEVIEYMN 300
301 DVKVLGPREL RVLLRWRKSM LETLEEQRKA VEGEAKEVEI EENLTEEQLE DRAMAEIDEL 360
361 IAKASEDEKA ALKKKKKKML KAKARVLKRR ELKMIIDGDE GPQAEDQEVF QLKKIRKAKE 420
421 LAEITKDTQA PDFDMMEDGD SDEEEGLGEG EWETHGEEGE NSDEDENELI HTEKSGMSKM 480
481 EKRNARTESW FEKEEISGLV SDEDDDDEMN AIEKHIGKNR SKKTENYENT VSFDDSKGKK 540
541 KGKKGGKGGE DDGFNTNDAD EQASESESSD AEMDEVAKEK LSRFDAQIDL DDDDEEERYE 600
601 DEGRRAAKRK CDKKIIGEDL KPVAKKRKLT PEQLAIGEQM IYSAKAARNL EDEAWNRYAN 660
661 NDEDLPDWFA DDEKKHYFKQ TPVTKEQVAL YRERMREFNA RPSKKVAEAK ARKQKKMQRK 720
721 LESAKKKAEG ILENDQMEHS EKVREMKKVY ANATRKEKKK VELVRMTKGK KGKTGRPNGQ 780
781 YKLVDR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
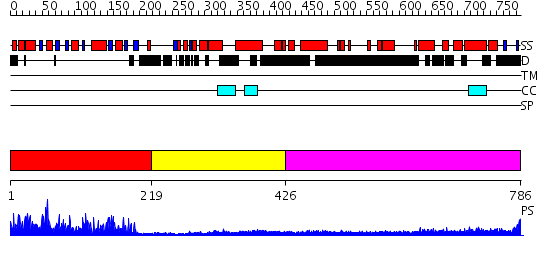
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..218] | 77.0 | RNA methyltransferase FtsJ | |
| 2 | View Details | [219..425] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [426..786] | 13.69897 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)