













| General Information: |
|
| Name(s) found: |
ahr-1 /
CE38293
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 560 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA intracellular [TAS |
| Biological Process: |
signal transduction
[IEA regulation of transcription, DNA-dependent [IEA response to xenobiotic stimulus [IDA regulation of transcription [IEA intracellular receptor mediated signaling pathway [TAS |
| Molecular Function: |
DNA binding
[IDA transcription regulator activity [IEA signal transducer activity [IEA aryl hydrocarbon receptor activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLLPYDSSTI SRLDKLSVLR LAVSFLQCKA HFQACLHNSQ FLSAGFPMST HSYSYQPHPP 60
61 IPFSNKVPTI FDLRIGTPML DPEESNFEEI SLKSLGGFIL VLNDNGEIYY ASENVENYLG 120
121 FHQSDVLHQP VYDLIHSEDR DDIRQQLDSN FHIPTSSASN QFDVFAPQNS KYLERNVNAR 180
181 FRCLLDNTCG FLRIDMRGKL MSLHGLPSSY VMGRTASGPV LGMICVCTPF VPPSTSDLAS 240
241 EDMILKTKHQ LDGALVSMDQ KVYEMLEIDE TDLPMPLYNL VHVEDAVCMA EAHKEAIKNG 300
301 SSGLLVYRLV STKTRRTYFV QSSCRMFYKN SKPESIGLTH RLLNEVEGTM LLEKRSTLKA 360
361 KLLSFDDSFL QSPRNLQSTA ALPLPSVLKD DQDCLEPSTS NSLFPSVPVP TPTTTKANRR 420
421 RKENSHEIVP TIPSIPIPTH FDMQMFDPSW NHGVHPPAWP HDVYHLTQYP PTYPHPPGTV 480
481 GYPDVQIAPV DYPGWHPNDI HMTQLPHGFT PDAQKLVPPH PQMSHFTEYP TPSTHHDLHH 540
541 HPLKQDNFHL ISEVTNLLGT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
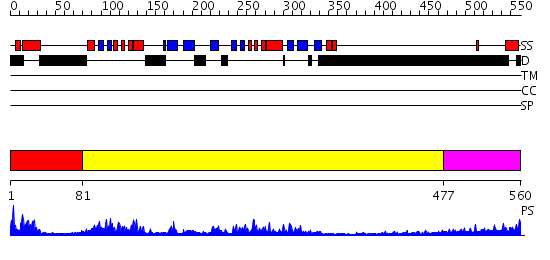
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..476] | 55.045757 | Crystal Structure of the PAS repeat region of the Drosophila clock protein PERIOD | |
| 3 | View Details | [477..560] | 5.254997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)