













| General Information: |
|
| Name(s) found: |
uba-2 /
CE40719
[WormBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 582 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
positive regulation of vulval development
[IMP hermaphrodite genitalia development [IMP embryonic development ending in birth or egg hatching [IMP positive regulation of growth rate [IMP protein modification process [IEA gamete generation [IMP ubiquitin cycle [IEA Mo-molybdopterin cofactor biosynthetic process [IEA growth [IMP reproduction [IMP locomotory behavior [IMP nematode larval development [IMP oviposition [IMP |
| Molecular Function: |
catalytic activity
[IEA small protein activating enzyme activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPSWREKHEK IVQSKILVIG AGGIGCELLK NLAVTGFRKV HVIDLDTIDI SNLNRQFLFR 60
61 KEHVSSSKAA TATQVVKQFC PQIELTFDHD SIFEKKYNME FFQAYDIVLN ALDNRAARNY 120
121 VNRMCHAANR PLIDSGSGGY FGQVSVIMRG KTECYECVDK PVQQTTYPGC TIRNTPSEHI 180
181 HCTVWAKHVF NQLFGEVDID DDVSPDMDAV DPDNTEAVTT EKEKEAMKEE PAPVGTRQWA 240
241 ESVDYDAAKV FDKLFLHDIE YLCKMEHLWK QRKRPSPLEF HTASSTGGEP QSLCDAQRDD 300
301 TSIWTLSTCA KVFSTCIQEL LEQIRAEPDV KLAFDKDHAI IMSFVAACAN IRAKIFGIPM 360
361 KSQFDIKAMA GNIIPAIAST NAIVAGIIVT EAVRVIEGST VICNSSIATT QSNPRGRIFG 420
421 GDATNPPNPR CFVCSEKREV FIYVNPDTMT VGGLCEKVLK QKLNMLAPDV MDSATSRIIV 480
481 SSDGDTDDLL PKKLAEVSIE DGAILSCDDF QQEMEIKLFI KKGDRLAGDD FEVARSEKEP 540
541 EPDDRKRKAD GSEEPEAKRQ KVEEKDDKNG NEAVAEITET MA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
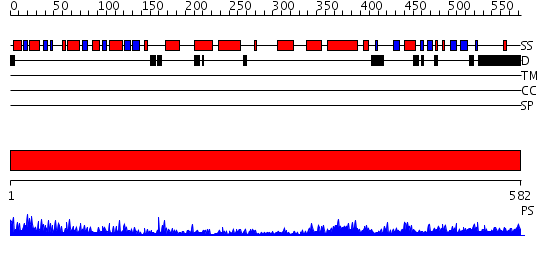
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..582] | 135.0 | SUMO E1 ACTIVATING ENZYME SAE1-SAE2-MG-ATP COMPLEX |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)