













| General Information: |
|
| Name(s) found: |
PLDG2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 856 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
cytosolic ribosome [IDA] |
| Biological Process: |
phospholipid catabolic process
[TAS]
|
| Molecular Function: |
phospholipase D activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSMGGGSNHE FGQWLDQQLV PLATSSGSLM VELLHGNLDI WVKEAKHLPN MICYRNKLVG 60
61 GISFSELGRR IRKVDGEKSS KFTSDPYVTV SISGAVIGRT FVISNSENPV WMQHFDVPVA 120
121 HSAAEVHFVV KDNDPIGSKI IGVVGIPTKQ LCSGNRIEGL FPILNSSGKP CRKGAMLSLS 180
181 IQYTPMERMR LYQKGVGSGV ECVGVPGTYF PLRKGGRVTL YQDAHVDDGT LPSVHLDGGI 240
241 QYRHGKCWED MADAIRRARR LIYITGWSVF HPVRLVRRNN DPTEGTLGEL LKVKSQEGVR 300
301 VLVLVWDDPT SMSFPGFSTK GLMNTSDEET RRFFKHSSVQ VLLCPRYGGK GHSFIKKSEV 360
361 ETIYTHHQKT MIVDAEAAQN RRKIVAFVGG LDLCNGRFDT PKHSLFGTLK TLHKDDFHNP 420
421 NFVTTEDVGP REPWHDLHSK IDGPAAYDVL ANFEERWMAS KPRGIGKGRT SFDDSLLRIN 480
481 RIPDIMGLSE ASSANDNDPE SWHVQVFRSI DSTSVKGFPK DPEEATGRNL LCGKNILIDM 540
541 SIHAAYVKAI RSAQHFIYIE NQYFLGSSFN WDSNKDLGAN NLIPMEIALK IANKIRAREN 600
601 FAAYIVIPMW PEGAPTSKPI QRILYWQHKT MQMMYQTIYK ALLEVGLDGQ LEPQDFLNFF 660
661 CLGNREVGTR EVPDGTVNVY NCPRKPPQPN AAQVQALKSR RFMIYVHSKG MVVDDEFVLI 720
721 GSANINQRSL EGTRDTEIAM GGYQPHHSWA KKGSRPRGQI FGYRMSLWAE HLGFLEQEFE 780
781 EPENMECVRR VRQLSELNWG QYAAEEVTEM SGHLLKYPVQ VDKTGKVSSL PGCETFPDLG 840
841 GKIIGSFLTL QENLTI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
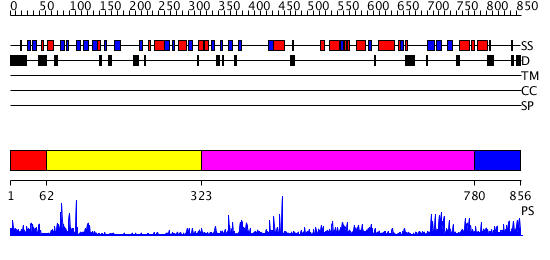
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..61] | 7.69897 | No description for 2eliA was found. | |
| 2 | View Details | [62..322] | 18.522879 | Synaptogamin I | |
| 3 | View Details | [323..779] | 7.154902 | Phospholipase D | |
| 4 | View Details | [780..856] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)