













| General Information: |
|
| Name(s) found: |
PARG2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 522 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
defense response to fungus
[IMP]
|
| Molecular Function: |
poly(ADP-ribose) glycohydrolase activity
[IEA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MELRADLRSI LQYLPLVAQS SSLVWPPSVE EELQTISRGP SESMVNSGEA LALHITNMRK 60
61 SLSLNASDLA PYALQGYGLF FDKKISREES ANFFGEVVPA LCRLLLQLPS MLEKHYQKAD 120
121 HVLDGVKSGL RLLGPQEAGI VLLSQELIAA LLACSFFCLF PEVDRSLKNL QGINFSGLFS 180
181 FPYMRHCTKQ ENKIKCLIHY FGRICRWMPT GFVSFERKIL PLEYHPHFVS YPKADSWANS 240
241 VTPLCSIEIH TSGAIEDQPC EALEVDFADE YFGGLTLSYD TLQEEIRFVI NPELIAGMIF 300
301 LPRMDANEAI EIVGVERFSG YTGYGPSFQY AGDYTDNKDL DIFRRRKTRV IAIDAMPDPG 360
361 MGQYKLDALI REVNKAFSGY MHQCKYNIDV KHDPEASSSH VPLTSDSASQ VIESSHRWCI 420
421 DHEEKKIGVA TGNWGCGVFG GDPELKIMLQ WLAISQSGRP FMSYYTFGLQ ALQNLNQVIE 480
481 MVALQEMTVG DLWKKLVEYS SERLSRRTWL GFFSWLMTSL ST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
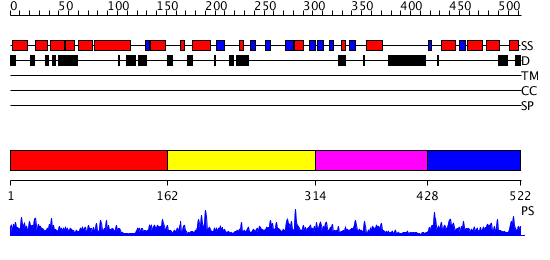
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [162..313] | 1.058959 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [314..427] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [428..522] | 9.068997 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)