













| General Information: |
|
| Name(s) found: |
ERF93_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 243 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
regulation of transcription, DNA-dependent
[TAS]
ethylene mediated signaling pathway [TAS] response to chitin [IEP] |
| Molecular Function: |
DNA binding
[TAS]
transcription factor activity [ISS][TAS] transcription activator activity [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEYSQSSMYS SPSSWSSSQE SLLWNESCFL DQSSEPQAFF CPNYDYSDDF FSFESPEMMI 60
61 KEEIQNGDVS NSEEEEKVGI DEERSYRGVR KRPWGKFAAE IRDSTRNGIR VWLGTFDKAE 120
121 EAALAYDQAA FATKGSLATL NFPVEVVRES LKKMENVNLH DGGSPVMALK RKHSLRNRPR 180
181 GKKRSSSSSS SSSNSSSCSS SSSTSSTSRS SSKQSVVKQE SGTLVVFEDL GAEYLEQLLM 240
241 SSC |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
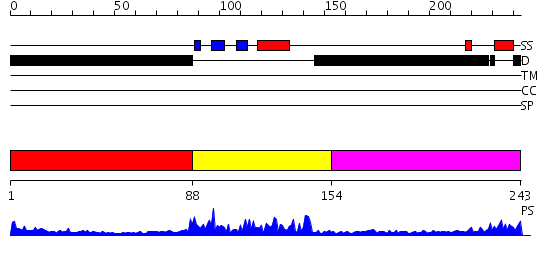
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [88..153] | 25.30103 | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | |
| 3 | View Details | [154..243] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)