













| General Information: |
|
| Name(s) found: |
hsdM /
EG10458
[EcoGene]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli K-12 |
| Length: | 529 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNNNDLVAKL WKLCDNLRDG GVSYQNYVNE LASLLFLKMC KETGQEAEYL PEGYRWDDLK 60
61 SRIGQEQLQF YRKMLVHLGE DDKKLVQAVF HNVSTTITEP KQITALVSNM DSLDWYNGAH 120
121 GKSRDDFGDM YEGLLQKNAN ETKSGAGQYF TPRPLIKTII HLLKPQPREV VQDPAAGTAG 180
181 FLIEADRYVK SQTNDLDDLD GDTQDFQIHR AFIGLELVPG TRRLALMNCL LHDIEGNLDH 240
241 GGAIRLGNTL GSDGENLPKA HIVATNPPFG SAAGTNITRT FVHPTSNKQL CFMQHIIETL 300
301 HPGGRAAVVV PDNVLFEGGK GTDIRRDLMD KCHLHTILRL PTGIFYAQGV KTNVLFFTKG 360
361 TVANPNQDKN CTDDVWVYDL RTNMPSFGKR TPFTDEHLQP FERVYGEDPH GLSPRTEGEW 420
421 SFNAEETEVA DSEENKNTDQ HLATSRWRKF SREWIRTAKS DSLDISWLKD KDSIDADSLP 480
481 EPDVLAAEAM GELVQALSEL DALMRELGAS DEADLQRQLL EEAFGGVKE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
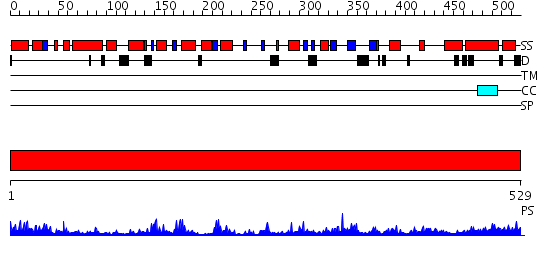
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..529] | 110.0 | Crystal structure of Type I restriction enzyme EcoKI M protein (EC 2.1.1.72) (M.EcoKI) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)