













| General Information: |
|
| Name(s) found: |
PARP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 983 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][ISS]
intracellular [IEA] |
| Biological Process: |
DNA repair
[IEP]
response to oxidative stress [IEP] response to abiotic stimulus [IMP] response to abscisic acid stimulus [IEP] protein amino acid ADP-ribosylation [ISS] |
| Molecular Function: |
DNA binding
[IEA][ISS]
zinc ion binding [IEA] NAD or NADH binding [IEA] NAD+ ADP-ribosyltransferase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASPHKPWRA EYAKSSRSSC KTCKSVINKE NFRLGKLVQS THFDGIMPMW NHASCILKKT 60
61 KQIKSVDDVE GIESLRWEDQ QKIRKYVESG AGSNTSTSTG TSTSSTANNA KLEYGIEVSQ 120
121 TSRAGCRKCS EKILKGEVRI FSKPEGPGNK GLMWHHAKCF LEMSSSTELE SLSGWRSIPD 180
181 SDQEALLPLV KKALPAAKTE TAEARQTNSR AGTKRKNDSV DNEKSKLAKS SFDMSTSGAL 240
241 QPCSKEKEME AQTKELWDLK DDLKKYVTSA ELREMLEVNE QSTRGSELDL RDKCADGMMF 300
301 GPLALCPMCS GHLSFSGGLY RCHGYISEWS KCSHSTLDPD RIKGKWKIPD ETENQFLLKW 360
361 NKSQKSVKPK RILRPVLSGE TSQGQGSKDA TDSSRSERLA DLKVSIAGNT KERQPWKKRI 420
421 EEAGAEFHAN VKKGTSCLVV CGLTDIRDAE MRKARRMKVA IVREDYLVDC FKKQRKLPFD 480
481 KYKIEDTSES LVTVKVKGRS AVHEASGLQE HCHILEDGNS IYNTTLSMSD LSTGINSYYI 540
541 LQIIQEDKGS DCYVFRKWGR VGNEKIGGNK VEEMSKSDAV HEFKRLFLEK TGNTWESWEQ 600
601 KTNFQKQPGK FLPLDIDYGV NKQVAKKEPF QTSSNLAPSL IELMKMLFDV ETYRSAMMEF 660
661 EINMSEMPLG KLSKHNIQKG FEALTEIQRL LTESDPQPTM KESLLVDASN RFFTMIPSIH 720
721 PHIIRDEDDF KSKVKMLEAL QDIEIASRIV GFDVDSTESL DDKYKKLHCD ISPLPHDSED 780
781 YRLIEKYLNT THAPTHTEWS LELEEVFALE REGEFDKYAP HREKLGNKML LWHGSRLTNF 840
841 VGILNQGLRI APPEAPATGY MFGKGIYFAD LVSKSAQYCY TCKKNPVGLM LLSEVALGEI 900
901 HELTKAKYMD KPPRGKHSTK GLGKKVPQDS EFAKWRGDVT VPCGKPVSSK VKASELMYNE 960
961 YIVYDTAQVK LQFLLKVRFK HKR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
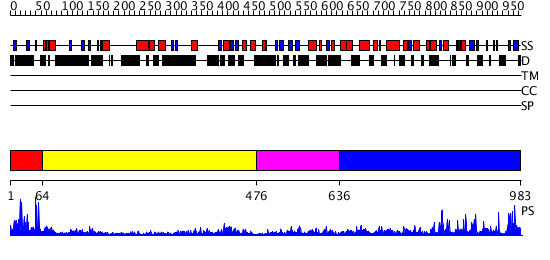
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..63] | 22.69897 | Solution structure of the first Zn-finger domain of poly(ADP-ribose) polymerase-1 | |
| 2 | View Details | [64..475] | 50.522879 | No description for 2owoA was found. | |
| 3 | View Details | [476..635] | 21.0 | Solution structure of WGR domain of poly(ADP-ribose) polymerase-1 | |
| 4 | View Details | [636..983] | 104.0 | Crystal structure of catalytic domain of human poly(ADP-ribose) polymerase with a novel inhibitor |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)