













| General Information: |
|
| Name(s) found: |
GLYG_RABIT
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Oryctolagus cuniculus |
| Length: | 333 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTDQAFVTLT TNDAYAKGAL VLGSSLKQHR TSRRLAVLTT PQVSDTMRKA LEIVFDEVIT 60
61 VDILDSGDSA HLTLMKRPEL GVTLTKLHCW SLTQYSKCVF MDADTLVLAN IDDLFEREEL 120
121 SAAPDPGWPD CFNSGVFVYQ PSVETYNQLL HVASEQGSFD GGDQGLLNTF FNSWATTDIR 180
181 KHLPFIYNLS SISIYSYLPA FKAFGANAKV VHFLGQTKPW NYTYDTKTKS VRSEGHDPTM 240
241 THPQFLNVWW DIFTTSVVPL LQQFGLVQDT CSYQHVEDVS GAVSHLSLGE TPATTQPFVS 300
301 SEERKERWEQ GQADYMGADS FDNIKKKLDT YLQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
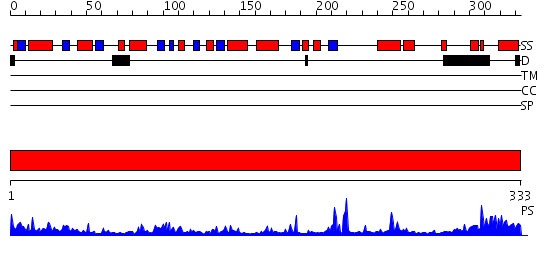
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..333] | 101.0 | apo form of the 162S mutant of glycogenin |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)