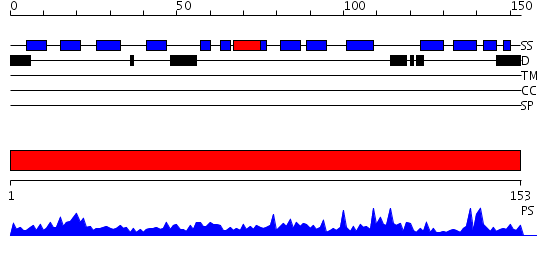
Description(s) found:
SHOW ONLY BEST
|
gi|88192069|pdb|2AIR|H Chain H, T-State Active Site Of Aspartate Transcarbamylase:crystal Structure Of The Carbamyl Phosphate And L-Alanosine Ligated Enzyme
[NCBI NR]
gi|88192067|pdb|2AIR|B Chain B, T-State Active Site Of Aspartate Transcarbamylase:crystal Structure Of The Carbamyl Phosphate And L-Alanosine Ligated Enzyme
[NCBI NR]
gi|78101181|pdb|2A0F|D Chain D, Structure Of D236a Mutant E. Coli Aspartate Transcarbamoylase In Presence Of Phosphonoacetamide At 2.90 A Resolution
[NCBI NR]
gi|78101179|pdb|2A0F|B Chain B, Structure Of D236a Mutant E. Coli Aspartate Transcarbamoylase In Presence Of Phosphonoacetamide At 2.90 A Resolution
[NCBI NR]
gi|71042237|pdb|1ZA2|D Chain D, Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In The Presence Of Ctp, Carbamoyl Phosphate At 2.50 A Resolution
[NCBI NR]
gi|71042235|pdb|1ZA2|B Chain B, Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In The Presence Of Ctp, Carbamoyl Phosphate At 2.50 A Resolution
[NCBI NR]
gi|71042233|pdb|1ZA1|D Chain D, Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In The Presence Of Ctp At 2.20 A Resolution
[NCBI NR]
gi|71042231|pdb|1ZA1|B Chain B, Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In The Presence Of Ctp At 2.20 A Resolution
[NCBI NR]
gi|6980789|pdb|1D09|D Chain D, Aspartate Transcarbamoylase Complexed With N-Phosphonacetyl- L-Aspartate (Pala)
[NCBI NR]
gi|6980787|pdb|1D09|B Chain B, Aspartate Transcarbamoylase Complexed With N-Phosphonacetyl- L-Aspartate (Pala)
[NCBI NR]
gi|67471965|sp|P0A7F3.2|PYRI_ECOLI RecName: Full=Aspartate carbamoyltransferase regulatory chain
[NCBI NR]
gi|67463873|pdb|1XJW|D Chain D, The Structure Of E. Coli Aspartate Transcarbamoylase Q137a Mutant In The R-State
[NCBI NR]
gi|67463871|pdb|1XJW|B Chain B, The Structure Of E. Coli Aspartate Transcarbamoylase Q137a Mutant In The R-State
[NCBI NR]
pir||DTECR aspartate carbamoyltransferase (EC 2.1.3.2) regulatory chain - Escherichia coli (strain K-12)�
[NCBI NR]
gi|537086|gb|AAA97141.1| aspartate carbomoyltransferase regulatory subunit [Escherichia coli]
[NCBI NR]
gi|51247749|pdb|1TUG|D Chain D, Aspartate Transcarbamoylase Catalytic Chain Mutant E50a Complex With Phosphonoacetamide, Malonate, And Cytidine-5- Prime-Triphosphate (Ctp)
[NCBI NR]
gi|51247747|pdb|1TUG|B Chain B, Aspartate Transcarbamoylase Catalytic Chain Mutant E50a Complex With Phosphonoacetamide, Malonate, And Cytidine-5- Prime-Triphosphate (Ctp)
[NCBI NR]
gi|51247741|pdb|1TU0|D Chain D, Aspartate Transcarbamoylase Catalytic Chain Mutant E50a Complex With Phosphonoacetamide
[NCBI NR]
gi|51247739|pdb|1TU0|B Chain B, Aspartate Transcarbamoylase Catalytic Chain Mutant E50a Complex With Phosphonoacetamide
[NCBI NR]
gi|51247735|pdb|1TTH|D Chain D, Aspartate Transcarbamoylase Catalytic Chain Mutant Glu50ala Complexed With N-(Phosphonacetyl-L-Aspartate) (Pala)
[NCBI NR]
gi|51247733|pdb|1TTH|B Chain B, Aspartate Transcarbamoylase Catalytic Chain Mutant Glu50ala Complexed With N-(Phosphonacetyl-L-Aspartate) (Pala)
[NCBI NR]
gi|49258620|pdb|1R0C|H Chain H, Products In The T State Of Aspartate Transcarbamylase: Crystal Structure Of The Phosphate And N-Carbamyl-L- Aspartate Ligated Enzyme
[NCBI NR]
gi|49258618|pdb|1R0C|B Chain B, Products In The T State Of Aspartate Transcarbamylase: Crystal Structure Of The Phosphate And N-Carbamyl-L- Aspartate Ligated Enzyme
[NCBI NR]
gi|49258616|pdb|1R0B|L Chain L, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[NCBI NR]
gi|49258615|pdb|1R0B|K Chain K, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[NCBI NR]
gi|49258614|pdb|1R0B|J Chain J, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[NCBI NR]
gi|49258610|pdb|1R0B|I Chain I, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[NCBI NR]
gi|49258609|pdb|1R0B|H Chain H, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[NCBI NR]
gi|49258608|pdb|1R0B|G Chain G, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[NCBI NR]
gi|49258581|pdb|1Q95|L Chain L, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[NCBI NR]
gi|49258580|pdb|1Q95|K Chain K, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[NCBI NR]
gi|49258579|pdb|1Q95|J Chain J, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[NCBI NR]
gi|49258578|pdb|1Q95|I Chain I, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[NCBI NR]
gi|49258577|pdb|1Q95|H Chain H, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[NCBI NR]
gi|49258576|pdb|1Q95|G Chain G, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[NCBI NR]
gi|47169159|pdb|1SKU|D Chain D, E. Coli Aspartate Transcarbamylase 240's Loop Mutant (K244n)
[NCBI NR]
gi|47169157|pdb|1SKU|B Chain B, E. Coli Aspartate Transcarbamylase 240's Loop Mutant (K244n)
[NCBI NR]
pir||F86122 aspartate carbamoyltransferase (EC 2.1.3.2) regulatory chain - Escherichia coli (strain O157:H7, substrain EDL933)�
[NCBI NR]
pir||E91281 aspartate carbamoyltransferase (EC 2.1.3.2) regulatory chain - Escherichia coli (strain O157:H7, substrain RIMD 0509952)�
[NCBI NR]
gi|209749616|gb|ACI73115.1| aspartate carbamoyltransferase regulatory subunit [Escherichia coli]
[NCBI NR]
gi|209749614|gb|ACI73114.1| aspartate carbamoyltransferase regulatory subunit [Escherichia coli]
[NCBI NR]
gi|209749612|gb|ACI73113.1| aspartate carbamoyltransferase regulatory subunit [Escherichia coli]
[NCBI NR]
gi|209749610|gb|ACI73112.1| aspartate carbamoyltransferase regulatory subunit [Escherichia coli]
[NCBI NR]
gi|209749608|gb|ACI73111.1| aspartate carbamoyltransferase regulatory subunit [Escherichia coli]
[NCBI NR]
gi|194709189|pdb|3D7S|D Chain D, Crystal Structure Of Wild-Type E. Coli Asparate Transcarbamoylase At Ph 8.5 At 2.80 A Resolution
[NCBI NR]
gi|194709187|pdb|3D7S|B Chain B, Crystal Structure Of Wild-Type E. Coli Asparate Transcarbamoylase At Ph 8.5 At 2.80 A Resolution
[NCBI NR]
gi|158428833|pdb|2IPO|D Chain D, E. Coli Aspartate Transcarbamoylase Complexed With N- Phosphonacetyl-L-Asparagine
[NCBI NR]
gi|158428831|pdb|2IPO|B Chain B, E. Coli Aspartate Transcarbamoylase Complexed With N- Phosphonacetyl-L-Asparagine
[NCBI NR]
gi|157879884|pdb|1RAI|D Chain D, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879883|pdb|1RAI|B Chain B, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879882|pdb|1RAH|D Chain D, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879881|pdb|1RAH|B Chain B, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879880|pdb|1RAG|D Chain D, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879879|pdb|1RAG|B Chain B, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879878|pdb|1RAF|D Chain D, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879877|pdb|1RAF|B Chain B, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879876|pdb|1RAE|D Chain D, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879875|pdb|1RAE|B Chain B, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879874|pdb|1RAD|D Chain D, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879873|pdb|1RAD|B Chain B, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879872|pdb|1RAC|D Chain D, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879871|pdb|1RAC|B Chain B, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879870|pdb|1RAB|D Chain D, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879869|pdb|1RAB|B Chain B, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879868|pdb|1RAA|D Chain D, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|157879867|pdb|1RAA|B Chain B, Crystal Structure Of Ctp-Ligated T State Aspartate Transcarbamoylase At 2.5 Angstroms Resolution: Implications For Atcase Mutants And The Mechanism Of Negative Cooperativity
[NCBI NR]
gi|152149096|pdb|2HSE|D Chain D, Structure Of D236a E. Coli Aspartate Transcarbamoylase In The Presence Of Phosphonoacetamide And L-Aspartate At 2.60 A Resolution
[NCBI NR]
gi|152149094|pdb|2HSE|B Chain B, Structure Of D236a E. Coli Aspartate Transcarbamoylase In The Presence Of Phosphonoacetamide And L-Aspartate At 2.60 A Resolution
[NCBI NR]
gi|147465|gb|AAA24477.1| aspartate transcarbamoylase regulatory chain (pyrI)
[NCBI NR]
gi|14278215|pdb|1I5O|D Chain D, Crystal Structure Of Mutant R105a Of E. Coli Aspartate Transcarbamoylase
[NCBI NR]
gi|14278213|pdb|1I5O|B Chain B, Crystal Structure Of Mutant R105a Of E. Coli Aspartate Transcarbamoylase
[NCBI NR]
gi|118137862|pdb|2H3E|D Chain D, Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In The Presence Of N-Phosphonacetyl-L-Isoasparagine At 2.3a Resolution
[NCBI NR]
gi|118137860|pdb|2H3E|B Chain B, Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In The Presence Of N-Phosphonacetyl-L-Isoasparagine At 2.3a Resolution
[NCBI NR]
gi|11513497|pdb|1F1B|D Chain D, Crystal Structure Of E. Coli Aspartate Transcarbamoylase P268a Mutant In The R-State In The Presence Of N- Phosphonacetyl-L-Aspartate
[NCBI NR]
gi|11513495|pdb|1F1B|B Chain B, Crystal Structure Of E. Coli Aspartate Transcarbamoylase P268a Mutant In The R-State In The Presence Of N- Phosphonacetyl-L-Aspartate
[NCBI NR]
gi|11513490|pdb|1EZZ|D Chain D, Crystal Structure Of E. Coli Aspartate Transcarbamoylase P268a Mutant In The T-State
[NCBI NR]
gi|11513488|pdb|1EZZ|B Chain B, Crystal Structure Of E. Coli Aspartate Transcarbamoylase P268a Mutant In The T-State
[NCBI NR]
gi|114793927|pdb|2FZK|D Chain D, The Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In Complex With Novel T State Inhibitors At 2.50 Resolution
[NCBI NR]
gi|114793925|pdb|2FZK|B Chain B, The Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In Complex With Novel T State Inhibitors At 2.50 Resolution
[NCBI NR]
gi|114793923|pdb|2FZG|D Chain D, The Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In Complex With Novel T State Inhibitors At 2.25 Resolution
[NCBI NR]
gi|114793921|pdb|2FZG|B Chain B, The Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In Complex With Novel T State Inhibitors At 2.25 Resolution
[NCBI NR]
gi|114793919|pdb|2FZC|D Chain D, The Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In Complex With Novel T State Inhibitors At 2.10 Resolution
[NCBI NR]
gi|114793917|pdb|2FZC|B Chain B, The Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In Complex With Novel T State Inhibitors At 2.10 Resolution
[NCBI NR]
Aspartate carbamoyltransferase regulatory chain - Escherichia coli
[Swiss-Prot]
aspartate carbamoyltransferase, regulatory subunit; aspartate carbamoyltransferase, regulatory subunit (allosteric regulation) [Escherichia coli K12]�gi|15834475|ref|NP_313248.1| aspartate carbamoyltransferase regulatory subunit [Escherichia coli O157:H7]
[nrdb_11152004_Chlamydomonas]
Aspartate carbamoyltransferase regulatory chain
[NCBI-RefSeq_human_na_08-09-2006_reversed.fasta]
aspartate carbomoyltransferase regulatory subunit [Escherichia coli]
[NCBI-RefSeq_human_na_08-09-2006_reversed.fasta]
Chain D, Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In The Presence Of Ctp, Carbamoyl Phosphate At 2.50 A Resolution
[nr-271106-contam.fasta]
Chain K, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[nr-271106-contam.fasta]
Chain B, The Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In Complex With Novel T State Inhibitors At 2.50 Resolution
[nr-271106-contam.fasta]
Chain D, Aspartate Transcarbamoylase Catalytic Chain Mutant E50a Complex With Phosphonoacetamide, Malonate, And Cytidine-5- Prime-Triphosphate (Ctp)
[nr-271106-contam.fasta]
Chain L, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[nr-271106-contam.fasta]
Chain B, Products In The T State Of Aspartate Transcarbamylase: Crystal Structure Of The Phosphate And N-Carbamyl-L- Aspartate Ligated Enzyme
[nr-271106-contam.fasta]
Chain D, Aspartate Transcarbamoylase Catalytic Chain Mutant Glu50ala Complexed With N-(Phosphonacetyl-L-Aspartate) (Pala)
[nr-271106-contam.fasta]
Chain B, Crystal Structure Of E. Coli Aspartate Transcarbamoylase P268a Mutant In The T-State
[nr-271106-contam.fasta]
Chain H, T-State Active Site Of Aspartate Transcarbamylase:crystal Structure Of The Carbamyl Phosphate And L-Alanosine Ligated Enzyme
[nr-271106-contam.fasta]
Chain D, Structure Of D236a Mutant E. Coli Aspartate Transcarbamoylase In Presence Of Phosphonoacetamide At 2.90 A Resolution
[nr-271106-contam.fasta]
Chain B, Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In The Presence Of Ctp At 2.20 A Resolution
[nr-271106-contam.fasta]
Chain B, Structure Of D236a Mutant E. Coli Aspartate Transcarbamoylase In Presence Of Phosphonoacetamide At 2.90 A Resolution
[nr-271106-contam.fasta]
aspartate transcarbamoylase regulatory chain (pyrI)
[nr-271106-contam.fasta]
Chain D, Crystal Structure Of E. Coli Aspartate Transcarbamoylase P268a Mutant In The T-State
[nr-271106-contam.fasta]
Chain I, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[nr-271106-contam.fasta]
Chain B, Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In The Presence Of N-Phosphonacetyl-L-Isoasparagine At 2.3a Resolution
[nr-271106-contam.fasta]
Chain D, E. Coli Aspartate Transcarbamylase 240's Loop Mutant (K244n)
[nr-271106-contam.fasta]
Chain B, Aspartate Transcarbamoylase Catalytic Chain Mutant E50a Complex With Phosphonoacetamide
[nr-271106-contam.fasta]
Chain G, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[nr-271106-contam.fasta]
Chain D, Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In The Presence Of N-Phosphonacetyl-L-Isoasparagine At 2.3a Resolution
[nr-271106-contam.fasta]
Chain D, Aspartate Transcarbamoylase Catalytic Chain Mutant E50a Complex With Phosphonoacetamide
[nr-271106-contam.fasta]
Chain J, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[nr-271106-contam.fasta]
Chain B, E. Coli Aspartate Transcarbamylase 240's Loop Mutant (K244n)
[nr-271106-contam.fasta]
Chain B, T-State Active Site Of Aspartate Transcarbamylase:crystal Structure Of The Carbamyl Phosphate And L-Alanosine Ligated Enzyme
[nr-271106-contam.fasta]
Chain D, Crystal Structure Of E. Coli Aspartate Transcarbamoylase P268a Mutant In The R-State In The Presence Of N- Phosphonacetyl-L-Aspartate
[nr-271106-contam.fasta]
Chain D, Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In The Presence Of Ctp At 2.20 A Resolution
[nr-271106-contam.fasta]
Chain D, The Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In Complex With Novel T State Inhibitors At 2.25 Resolution
[nr-271106-contam.fasta]
Chain B, Crystal Structure Of E. Coli Aspartate Transcarbamoylase P268a Mutant In The R-State In The Presence Of N- Phosphonacetyl-L-Aspartate
[nr-271106-contam.fasta]
Chain H, Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
[nr-271106-contam.fasta]
Chain B, The Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In Complex With Novel T State Inhibitors At 2.25 Resolution
[nr-271106-contam.fasta]
Chain D, The Structure Of E. Coli Aspartate Transcarbamoylase Q137a Mutant In The R-State
[nr-271106-contam.fasta]
Chain B, The Structure Of E. Coli Aspartate Transcarbamoylase Q137a Mutant In The R-State
[nr-271106-contam.fasta]
Chain D, Crystal Structure Of Mutant R105a Of E. Coli Aspartate Transcarbamoylase
[nr-271106-contam.fasta]
Chain B, Aspartate Transcarbamoylase Complexed With N-Phosphonacetyl- L-Aspartate (Pala)
[nr-271106-contam.fasta]
Chain B, Crystal Structure Of Mutant R105a Of E. Coli Aspartate Transcarbamoylase
[nr-271106-contam.fasta]
Chain D, The Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In Complex With Novel T State Inhibitors At 2.50 Resolution
[nr-271106-contam.fasta]
Chain D, Aspartate Transcarbamoylase Complexed With N-Phosphonacetyl- L-Aspartate (Pala)
[nr-271106-contam.fasta]
Chain B, Aspartate Transcarbamoylase Catalytic Chain Mutant E50a Complex With Phosphonoacetamide, Malonate, And Cytidine-5- Prime-Triphosphate (Ctp)
[nr-271106-contam.fasta]
Chain D, The Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In Complex With Novel T State Inhibitors At 2.10 Resolution
[nr-271106-contam.fasta]
Chain B, Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In The Presence Of Ctp, Carbamoyl Phosphate At 2.50 A Resolution
[nr-271106-contam.fasta]
Chain B, Aspartate Transcarbamoylase Catalytic Chain Mutant Glu50ala Complexed With N-(Phosphonacetyl-L-Aspartate) (Pala)
[nr-271106-contam.fasta]
Chain H, Products In The T State Of Aspartate Transcarbamylase: Crystal Structure Of The Phosphate And N-Carbamyl-L- Aspartate Ligated Enzyme
[nr-271106-contam.fasta]
Chain B, The Structure Of Wild-Type E. Coli Aspartate Transcarbamoylase In Complex With Novel T State Inhibitors At 2.10 Resolution
[nr-271106-contam.fasta]
Aspartate carbamoyltransferase regulatory chain OS=Escherichia coli (strain K12) GN=pyrI PE=1 SV=2
[uniprot-Ecoli-reviewed-110511.fasta]
|