













| General Information: |
|
| Name(s) found: |
TA12B_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 683 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
transcription factor TFIID complex [ISS] |
| Biological Process: |
regulation of ethylene mediated signaling pathway
[IMP]
transcription initiation [ISS] jasmonic acid mediated signaling pathway [IMP] |
| Molecular Function: |
transcription initiation factor activity
[IEA][ISS]
DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEPIPSSSL SPKSLQSPNP MEPSPASSTP LPSSSSQQQQ LMTAPISNSV NSAASPAMTV 60
61 TTTEGIVIQN NSQPNISSPN PTSSNPPIGA QIPSPSPLSH PSSSLDQQTQ TQQLVQQTQQ 120
121 LPQQQQQIMQ QISSSPIPQL SPQQQQILQQ QHMTSQQIPM SSYQIAQSLQ RSPSLSRLSQ 180
181 IQQQQQQQHQ GQYGNVLRQQ AGLYGTMNFG GSGSVQQSQQ NQQMVNPNMS RAGLVGQSGH 240
241 LPMLNGAAGA AQMNIQPQLL AASPRQKSGM VQGSQFHPGS SGQQLQGMQA MGMMGSLNLT 300
301 SQMRGNPALY AQQRINPGQM RQQLSQQNAL TSPQVQNLQR TSSLAFMNPQ LSGLAQNGQA 360
361 GMMQNSLSQQ QWLKQMSGIT SPNSFRLQPS QRQALLLQQQ QQQQQQLSSP QLHQSSMSLN 420
421 QQQISQIIQQ QQQQSQLGQS QMNQSHSQQQ LQQMQQQLQQ QPQQQMQQQQ QQQQQMQINQ 480
481 QQPSPRMLSH AGQKSVSLTG SQPEATQSGT TTPGGSSSQG TEATNQLLGK RKIQDLVSQV 540
541 DVHAKLDPDV EDLLLEVADD FIDSVTSFAC SLAKHRKSSV LEPKDILLHL EKNLHLTIPG 600
601 FSSEDKRQTK TVPTDLHKKR LAMVRALLES SKPETNASNS KETMRQAMVN PNGPNHLLRP 660
661 SQSSEQLVSQ TSGPHILQHM TRY |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
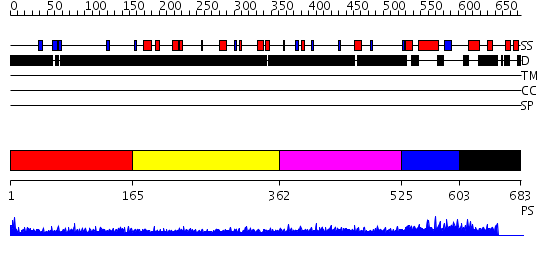
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..164] | 1.24 | No description for 2j63A was found. | |
| 2 | View Details | [165..361] | 4.0 | Sec24 | |
| 3 | View Details | [362..524] | 1.16 | Ribonuclease domain of colicin E3; Colicin E3 translocation domain; Colicin E3 receptor domain | |
| 4 | View Details | [525..602] | 34.221849 | TAF(II)-20, (TAF(II)-15, hTAF12), histone fold domain | |
| 5 | View Details | [603..683] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 |
|
||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)