













| General Information: |
|
| Name(s) found: |
gi|45443443
[NCBI NR]
gi|45438312 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Yersinia pestis biovar Microtus str. 91001 |
| Length: | 379 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKRDYYEVL GVSRDAEERE IKKAYKRLAM KFHPDRQSED KNAEEKFKEA KEAYEILTDA 60
61 QKRAAYDQYG HAAFEQGGMG GGGFGGGGGG ADFSDIFGDV FGDIFGGGRR QQRASRGSDL 120
121 RYNMDLTLEE AVRGVTKEIR IPTLDECDVC HGSGAKPGSS PVTCPTCHGA GQVQMRQGFF 180
181 TVQQACPHCH GRGQIIKDPC NKCHGHGRVE KSKTLSVKIP AGVDTGDRIR LSGEGEAGEH 240
241 GAPSGDLYVQ VQVKAHPIFE REGNNLYCEV PINFAMAALG GEIEVPTLDG RVKLKIPAET 300
301 QTGKMFRMRG KGVKSVRGGS QGDLLCRVVV ETPVSLSEKQ KQLLRELEES FVGAAGEKNS 360
361 PRAKSFLDGV KKFFDDLTR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
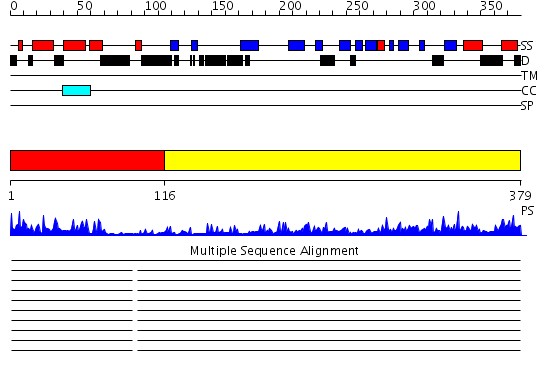
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | 25.045757 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [116..379] | 44.69897 | The crystal structure of Hsp40 Ydj1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)