













| General Information: |
|
| Name(s) found: |
gi|30065085
[NCBI NR]
gi|30043346 [NCBI NR] |
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Shigella flexneri 2a str. 2457T |
| Length: | 430 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKARQQKYCD KIANFWCHPT GKIIMSLAGK KIVLGVSGGI AAYKTPELVR RLRDRGADVR 60
61 VAMTEAAKAF ITPLSLQAVS GYPVSDSLLD PAAEAAMGHI ELGKWADLVI LAPATADLIA 120
121 RVAAGMANDL VSTICLATPA PVAVLPAMNQ QMYRAAATQH NLEVLASRGL LIWGPDSGSQ 180
181 ACGDIGPGRM LDPLTIVDMA VAHFSPVNDL KHLNIMITAG PTREPLDPVR YISNHSSGKM 240
241 GFAIAAAAAR RGANVTLVSG PVSLPTPPFV KRVDVMTALE MEAAVNASVQ QQNIFIGCAA 300
301 VADYRAATVA PEKIKKQATQ GDELTIKMVK NPDIVAGVAA LKDHRPYVVG FAAETNNVEE 360
361 YARQKRIRKN LDLICANDVS QPTQGFNSDN NALHLFWQDG DKVLPLERKE LLGQLLLDEI 420
421 VTRYDEKNRR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
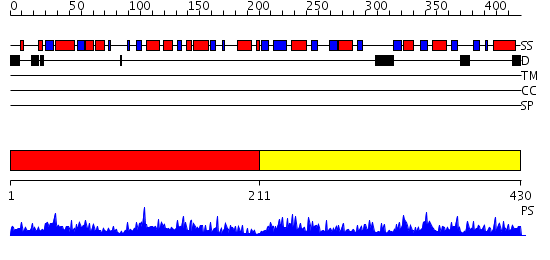
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..210] | 42.0 | crystal structure of human phosphopantothenoylcysteine decarboxylase | |
| 2 | View Details | [211..430] | 76.045757 | Phosphopantothenoylcysteine synthetase from E. coli |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)