













| General Information: |
|
| Name(s) found: |
RAPA_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 968 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPFTLGQRWI SDTESELGLG TVVAVDARTV TLLFPSTGEN RLYARSDSPV TRVMFNPGDT 60
61 ITSHDGWQMQ VEEVKEENGL LTYIGTRLDT EESGVALREV FLDSKLVFSK PQDRLFAGQI 120
121 DRMDRFALRY RARKYSSEQF RMPYSGLRGQ RTSLIPHQLN IAHDVGRRHA PRVLLADEVG 180
181 LGKTIEAGMI LHQQLLSGAA ERVLIIVPET LQHQWLVEML RRFNLRFALF DDERYAEAQH 240
241 DAYNPFDTEQ LVICSLDFAR RSKQRLEHLC EAEWDLLVVD EAHHLVWSED APSREYQAIE 300
301 QLAEHVPGVL LLTATPEQLG MESHFARLRL LDPNRFHDFA QFVEEQKNYR PVADAVAMLL 360
361 AGNKLSNDEL NMLGEMIGEQ DIEPLLQAAN SDSEDAQSAR QELVSMLMDR HGTSRVLFRN 420
421 TRNGVKGFPK RELHTIKLPL PTQYQTAIKV SGIMGARKSA EDRARDMLYP ERIYQEFEGD 480
481 NATWWNFDPR VEWLMGYLTS HRSQKVLVIC AKAATALQLE QVLREREGIR AAVFHEGMSI 540
541 IERDRAAAWF AEEDTGAQVL LCSEIGSEGR NFQFASHMVM FDLPFNPDLL EQRIGRLDRI 600
601 GQAHDIQIHV PYLEKTAQSV LVRWYHEGLD AFEHTCPTGR TIYDSVYNDL INYLASPDQT 660
661 EGFDDLIKNC REQHEALKAQ LEQGRDRLLE IHSNGGEKAQ ALAESIEEQD DDTNLIAFAM 720
721 NLFDIIGINQ DDRGDNMIVL TPSDHMLVPD FPGLSEDGIT ITFDREVALA REDAQFITWE 780
781 HPLIRNGLDL ILSGDTGSST ISLLKNKALP VGTLLVELIY VVEAQAPKQL QLNRFLPPTP 840
841 VRMLLDKNGN NLAAQVEFET FNRQLNAVNR HTGSKLVNAV QQDVHAILQL GEAQIEKSAR 900
901 ALIDAARNEA DEKLSAELSR LEALRAVNPN IRDDELTAIE SNRQQVMESL DQAGWRLDAL 960
961 RLIVVTHQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
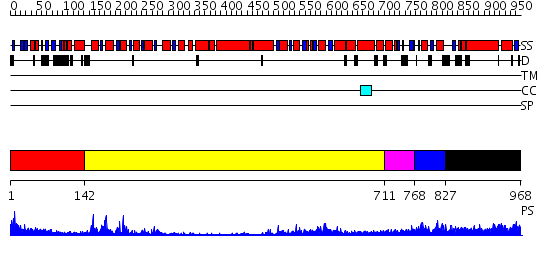
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..141] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [142..710] | 62.045757 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | |
| 3 | View Details | [711..767] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [768..826] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [827..968] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)