













| General Information: |
|
| Name(s) found: |
gi|50941323
[NCBI NR]
gi|37805865 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 725 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEINSSGEEA VVKVRKPYTI TKQRERWTEA EHNRFLEALK LYGRAWQRIE EHVGTKTAVQ 60
61 IRSHAQKFFT KLEKEAINNG TSPGQAHDID IPPPRPKRKP NSPYPRKSCL SSETSTREVQ 120
121 NDKATISNMT NNSTAQMAGD AALEKLTYIQ KLQRKEISEK GSCSEVLNLF REVPSASFSS 180
181 VNKSSSNHGA SRGLEPTKTE VKDVVILERD SISNGAGKDA KDINDQEMER LNGIHISSKP 240
241 DHSHENCLDT SSQQFKPKSN SVETTYVDWS AAKASHYQMD RNGVTGFQAT GTEGSHPDQT 300
301 SDQMGGASGT MNQCIHPTLP VDPKFDGNAA AQPFPHNYAA FAPMMQCHCN QDAYRSFANM 360
361 SSTFSSMLVS TLLSNPAIHA AARLAASYWP TVDGNTPDPN QENLSESAQG SHAGSPPNMA 420
421 SIVTATVAAA SAWWATQGLL PLFPPPIAFP FVPAPSAPFS TADVQRAQEK DIDCPMDNAQ 480
481 KELQETRKQD NFEAMKVIVS SETDESGKGE VSLHTELKIS PADKADTKPA AGAETSDVFG 540
541 NKKKQDRSSC GSNTPSSSDI EADNAPENQE KANDKAKQAS CSNSSAGDNN HRRFRSSAST 600
601 SDSWKEVSEE GRLAFDALFS RERLPQSFSP PQVEGSKEIS KEEEDEVTTV TVDLNKNAAI 660
661 IDQELDTADE PRASFPNELS NLKLKSRRTG FKPYKRCSVE AKENRVPASD EVGTKRIRLE 720
721 SEAST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
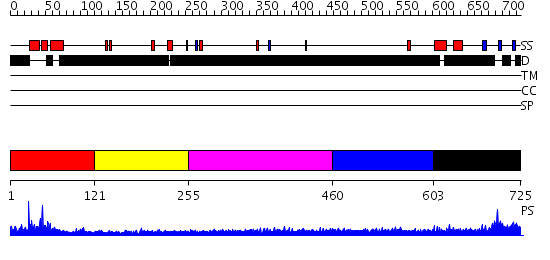
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 7.69897 | No description for 2v1dB was found. | |
| 2 | View Details | [121..254] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [255..459] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [460..602] | 3.213993 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [603..725] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)