













| General Information: |
|
| Name(s) found: |
ORR21_ORYSJ
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 691 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPVEDGGGV EFPVGMKVLV VDDDPTCLAV LKRMLLECRY DATTCSQATR ALTMLRENRR 60
61 GFDVIISDVH MPDMDGFRLL ELVGLEMDLP VIMMSADSRT DIVMKGIKHG ACDYLIKPVR 120
121 MEELKNIWQH VIRKKFNENK EHEHSGSLDD TDRTRPTNND NEYASSANDG AEGSWKSQKK 180
181 KRDKDDDDGE LESGDPSSTS KKPRVVWSVE LHQQFVNAVN HLGIDKAVPK KILELMNVPG 240
241 LTRENVASHL QKFRLYLKRI AQHHAGIANP FCPPASSGKV GSLGGLDFQA LAASGQIPPQ 300
301 ALAALQDELL GRPTNSLVLP GRDQSSLRLA AVKGNKPHGE REIAFGQPIY KCQNNAYGAF 360
361 PQSSPAVGGM PSFSAWPNNK LGMADSTGTL GGMSNSQNSN IVLHELQQQP DAMLSGTLHS 420
421 LDVKPSGIVM PSQSLNTFSA SEGLSPNQNT LMIPAQSSGF LAAMPPSMKH EPVLATSQPS 480
481 SSLLGGIDLV NQASTSQPLI SAHGGGNLSG LVNRNPNVVP SQGISTFHTP NNPYLVSPNS 540
541 MGMGSKQPPG VLKTENSDAL NHSYGYLGGS NPPMDSGLLS SQSKNTQFGL LGQDDITGSW 600
601 SPLPNVDSYG NTVGLSHPGS SSSSFQSSNV ALGKLPDQGR GKNHGFVGKG TCIPSRFAVD 660
661 EIESPTNNLS HSIGSSGDIM SPDIFGFSGQ M |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
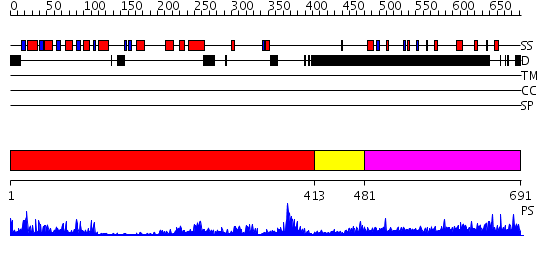
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..412] | 65.045757 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 2 | View Details | [413..480] | 53.69897 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding | |
| 3 | View Details | [481..691] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)