













| General Information: |
|
| Name(s) found: |
gi|37536642
[NCBI NR]
gi|31433391 [NCBI NR] gi|13876527 [NCBI NR] gi|115483276 [NCBI NR] gi|113639840 [NCBI NR] |
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 757 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATAPLAFHL PFPFPSASRP PPRLLPPSRR PPAARLAATR RFRPPTADDE PPEAAEDSSH 60
61 GLNRYDQLTR HVERARRRQQ AEQPEITPDH PLFSSPPSSG EAGSYDPDDE FFDEIDRAIA 120
121 EKREEFTRRG LIKPSAPAPS QPEEEDGLAD ELSPEEVIDL DEIRRLQGLS VVSLADEEDE 180
181 EANGGGGGVD YGDDGVPLDD DGEVFDVADE VGLEGARVRY PAFRMTLAEL LDESKLVPVA 240
241 VTGDQDVALA GVQRDASLVA AGDLYVCVGE EGLAGLTEAD KRGAVAVVAD QTVDIEGTLA 300
301 CRALVIVDDI TAALRMLPAC LYRRPSKDMA VIGVAGTDGV TTTAHLVRAM YEAMGVRTGM 360
361 VGVLGAYAFG NNKLDAQPDA SGDPIAVQRL MATMLYNGAE AALLEATTDG MPSSGVDSEI 420
421 DYDIAVLTNV RHAGDEAGMT YEEYMNSMAS LFSRMVDPER HRKVVNIDDP SAPFFAAQGG 480
481 QDVPVVTYSF ENKKADVHTL KYQLSLFETE VLVQTPHGIL EISSGLLGRD NIYNILASVA 540
541 VGVAVGAPLE DIVKGIEEVD AIPGRCELID EEQAFGVIVD HARTPESLSR LLDGVKELGP 600
601 RRIVTVIGCC GERERGKRPV MTKVAAEKSD VVMLTSDNPA NEDPLDILDD MLAGVGWTME 660
661 EYLKHGTNDY YPPLPNGHRI FLHDIRRVAV RAAVAMGEQG DVVVITGKGN DTYQIEVDKK 720
721 EFFDDREECR EALQYVDQLH RAGIDTSEFP WRLPESH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
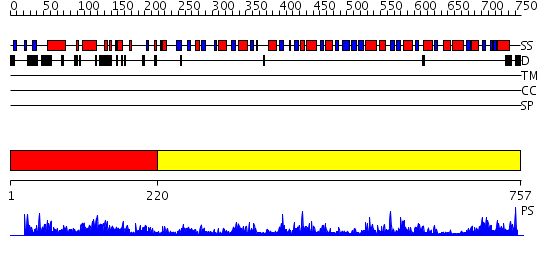
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..219] | 1.083995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [220..757] | 97.0 | UDP-N-acetylmuramyl tripeptide synthetase MurE |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.68 |
Source: Reynolds et al. (2008)