













| General Information: |
|
| Name(s) found: |
gi|50939471
[NCBI NR]
gi|33146722 [NCBI NR] gi|125601231 [NCBI NR] gi|115473603 [NCBI NR] gi|113611936 [NCBI NR] |
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 652 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEYATAKTS VWWDIENCQV PRACDPNLIA QNMSSALAAA GYTGPVSISA YGDIGRIGNA 60
61 VTHALSSTGI SLNHVPAGIK DASDKKILVD MLFWAIDNPP PANYLLISGD RDFSNALHKL 120
121 TMRRYNILLA QPPNVSQALT AAAKSVWLWK SLVAGEPPLA ESPYVSSTAS GNMVELDKSK 180
181 NINSDSSDTT TDTNPQNGLQ SDHQKGGNGK ADKQSKVKQP RRNQSDNVSK PASNEENSVE 240
241 VADNSKEYTT DHPTQSSMPS SSSSSSSESQ DGAKVNQSSK PKVQPFSLPK KPAKSAHCHQ 300
301 KTAPHDFNSK KSGASAESAA KNGTPDSGNG GGYNPKHHKP HTSQSPRPQN SITHPHSGSE 360
361 IFHHTLSSQR TNSCSPSAGH NGAPTAPLQS WPSAPPYHSP PVNYPDLNRI NISGYPRGIH 420
421 DNQGVNMNYH PNHSGSPHNV QPAYNSYRPP TPPSMPSNMQ NAGQWGVNPG YPQPSSDPQG 480
481 LIRNILGALE VLKTEKIPPI EQHISDCIRY GEANLPNFDV KKALELAIQH QAIVLKMLGP 540
541 MSFYLGKNQN LWKCVNIMDI NAKYPKDTFD AVHRFISSTS GSSAIKNSRS KYQAAIVLKN 600
601 QCLKHLALGE VLQILYIIIN TKKWFVPHSS GWQPLSFNII VVDATTGAGG KA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
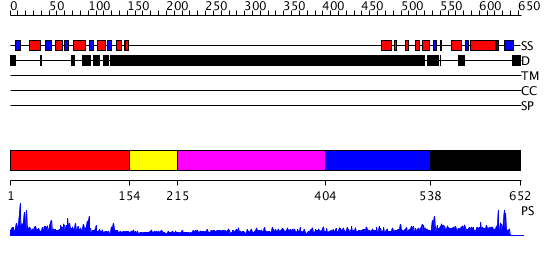
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..153] | 1.45 | No description for 2qipA was found. | |
| 2 | View Details | [154..214] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [215..403] | 1.266995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [404..537] | 4.267986 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [538..652] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)