













| General Information: |
|
| Name(s) found: |
gi|50926592
[NCBI NR]
gi|38344443 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 1989 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEQAKAHNL SPSVSGDDGE PNPRRRARTP PPPPRQSPRR EEALERVEGS ATSTSTGDGE 60
61 GRRDGERRLL VYGDGSTPQG ALLRHPPIVP DPESPAQRWL DDVANLVMTA RQRLGAGGRS 120
121 SATKTSGAAT TGSVSSRRRA RRAAAVARHS AATPSSAPPT QEDLRGGPDA RISIERQRNG 180
181 RRAAHATEGA SSSGVSHQHG RGDQPSVPPV GGVGCRAFVA SLRNVRWPPR FRPTIAEKYD 240
241 GSVNPAEFLQ VYTTGIEAAG GDDRVMANFF PMALKGQARG WLMNLPPASV HSWEDLCQQF 300
301 TMNFQGTYPH PGEEADLHAV QRRDDESLRS YIQRFCQVRN TIPCIPAHAV IYAFRGGVRH 360
361 NRMLEKIASK EPQTTAELFQ LADRVARKEE AWTWNPSGSG VAASAAPGSA ARTGRRDRRK 420
421 KKRSAHSDDE GHVLAVEGAS RATRKGRPAT DKKKEAGAPS RERPTGKWCI VHNTSLHDLT 480
481 DCRAVKSLAE RTRKWEEERR QERREGKSPA VPSGNRRSEA KQKAPAEDID DGDDDLGFQE 540
541 PGATIATVDG GACAHVSRRS LKAMKRELLA AAPTHEATRR ARWSEVALTF DQTDHPPCVA 600
601 RGGQIAMVVS PTVCNVKLGR VLIDGGAALN ILSPAAFDAI KAPGMVLRPS QPIIGVTPGH 660
661 TWPPGHIDLP VTFGGSANFR TERVNFDVAD LSLPYNAVLG RPALVKFMAA VHYAYLQMKM 720
721 PGPGGPISVH GDLKVALACM EQRADHLAAA SKPEGGDERL GTSVPAAPRQ RIVTCDEVPE 780
781 DALVSFLRAN ADVFAWRPAD MPGVPREVIE HRLAVRSGAR PVRQKVRRQA PERQAFIREE 840
841 VARLLEAGFI REVIHPEWLA NPVVVPKANG KLRMCIDYTD LNKACPKDPY PLPRIDQIVD 900
901 STAGCDLLCF LDAYSGYHQI RMAREDEEKT AFITPIGTYC YTTMPFGLKN AGPTFQRTTR 960
961 ISLGSQIGRN VEAYVDDLVV KTRNQETLLS DLAETFESLR SARIKLNPDK CVFGVPAGKL1020
1021 LGFLVSARGI EANPEKIRAI ERMRPPSKLR DVQCITGCMA ALSRFISRLG EKALPLFKLL1080
1081 KRSGPFTWTE EAERALTQLK AYLSSPPVLV ALEPNEPLLL YLAATPQVVS AALVVERDED1140
1141 NPHSAHPHPV LAWPGREQGG EAPEPNGGPR PPTTGAGPLP ACQTVPGAPD PQDGPRATAG1200
1201 RPHLSPSDPE ANPVPTRPGR EQGEEAPEPN GGLRPLTTGV GPLPACPTTP GAPDPQDGPE1260
1261 ATVGRPPLSS SDPEVVGTED ECTPEALRDA KTRYPQAQKM FYAILIASRK LRHYFQAHRV1320
1321 TVVTSYPLGQ ILHNREGTRR VVKWAIELSE FDLHFEPRHA IKSQALADFV AEWTPAPEPV1380
1381 SVPEASSGPS QPPHTAHWVM QFDGSLSLQG AGAGVTLTSP SGDVLRYLVR LDFQATNNMA1440
1441 EYEGLLAGLR VAAGLGIRRL LVLGDSQLVV NQVCKEYRCS DPQMDAYVRQ VRCMEHHFDG1500
1501 IELRHVPRRD NTVADELSRL ASSRAETPPG AFEERLAQPS ARPDPLGETD APDRPPRPVG1560
1561 VQASGPEGSA PSSLRLIAWI AEIQAYLTDK TLPEDREGSE RVQRISKRYV LVEGTLYRRA1620
1621 ANGILLKCIP REQGVELLAN IHEGECGAHS ASRTLVGKAF RQGFYWPTAL NDAVDLVRRC1680
1681 RACQFHAKQI HQPAQALQII PLSWSFAVWG LDILGPFRRA PGGFEYLYVA IDKFTKWPEA1740
1741 YPVVKIDKHS ALKFIKGITA RFGVPNRIIT DNGTQFTSEL FGDYCEDMGI KLCFASPAHP1800
1801 RSNGQVERAN AEILRGLKTK TFNILKKHGD SWIEELPAVL WANQTTPSRA TGETPFFLVY1860
1861 GAEAVLPSEL TLRSPRATIY CEADQDQLRR DDLDYLEERR RRAALRAARY QQSLRRYHQR1920
1921 HVRARSLCVD DLVLRRVQTR AGLSKLSPMW EGPYRVIGVP RPGSVRMATG DGTELPNPWN1980
1981 IEHLRRFYP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
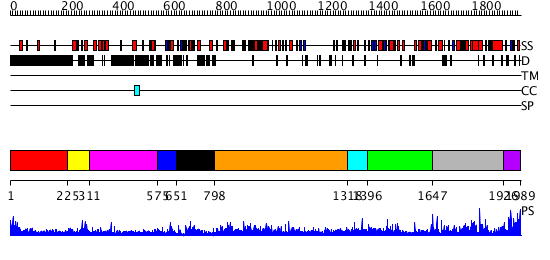
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..224] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [225..310] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [311..574] | 17.221849 | No description for 3dikA was found. | |
| 4 | View Details | [575..650] | 5.045757 | HIV nucleocapsid | |
| 5 | View Details | [651..797] | 26.09691 | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | |
| 6 | View Details | [798..1317] | 136.0 | No description for 3drsA was found. | |
| 7 | View Details | [1318..1395] | 6.69897 | N-terminal Zn binding domain of HIV integrase | |
| 8 | View Details | [1396..1646] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1647..1925] | 38.39794 | N-terminal Zn binding domain of HIV integrase; Retroviral integrase, catalytic domain | |
| 10 | View Details | [1926..1989] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)