













| General Information: |
|
| Name(s) found: |
gi|34896966
[NCBI NR]
gi|14018081 [NCBI NR] gi|125586578 [NCBI NR] gi|115453435 [NCBI NR] gi|113548789 [NCBI NR] gi|108708671 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 748 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MECNKDEALR AKEIAERKFE SKDLQGAKKF ALKAQALFPG LEGIVQMITT LDLYLASEVL 60
61 ISGEKDWYSI LSVESSADDE TLKKQYRKLV LQLHPDKNKS VGAEGAFKMV QEAWTVLSDK 120
121 TKRALYDQKR KLMVLKRNTS QTNKASAAPG ASNGFYNFAA NAAASKVTRG NKQKAGPATS 180
181 SVRQRPPPPP PPPRQAPAPP PAKPPTFWTS CNKCKMNYEY LKVYLNHNLL CPTCREPFLA 240
241 QEVPMPPTES VHAVHDPNIS GANQNTNGSR NFQWGPFSRT AGAASATASS AAAAQAANVV 300
301 HHTYEKVRRE REEAQAAARR EEALRRKYNP PKRQANISEN LNLGTGGNSS KKMRTMGNDI 360
361 GIGSSSILSG SGANYFGVPG GNISFSTNSG AHHFQGVNGG FSWKPRPPTR ISLVKTFTQF 420
421 DVRGILMEKA KSDLKDKLKE MQTKRSQVAA NGKKNKKNMF KESGGDDESL ASDDSTARQA 480
481 AHVDPEDNAS VNSTDADDEN DDPLSYNVPD PDFHDFDKDR TEECFQSDQI WATYDDEDGM 540
541 PRYYAFIQKV LSLEPFQLKI SFLTSRTNSE FGSLNWVSSG FTKTCGDFRI CRYETCDILN 600
601 MFSHQIKWEK GPRGVIKIYP QKGNIWAVYR NWSPDWDEDT PDKVLHAYDV VEVLDEYDED 660
661 LGISVIPLVK VAGFRTVFQR NQDLNAIKKI PKEEMFRFSH EVPFYRMSGE EAPNVPKDSY 720
721 ELDPAAISKE LLQEITETVE SSKATSEC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
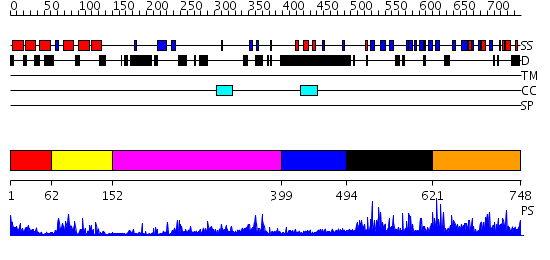
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..61] | 1.319994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [62..151] | 19.221849 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 3 | View Details | [152..398] | 35.69897 | The crystal structure of Hsp40 Ydj1 | |
| 4 | View Details | [399..493] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [494..620] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [621..748] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 |
|
||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. | ||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)