













| General Information: |
|
| Name(s) found: |
PLCD5_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 578 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
intracellular signaling pathway
[IEA]
signal transduction [IEA][ISS] lipid metabolic process [IEA] |
| Molecular Function: |
phosphoinositide phospholipase C activity
[IEA]
phospholipase C activity [IEA][ISS] calcium ion binding [IEA] phosphoric diester hydrolase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRDMGSYKM GLCCSDKLRM NRGAPPQDVV TAFVEYTEGR SHMTAEQLCR FLVEVQDETE 60
61 VLVSDAEKII ERITCERHHI TKFLRHTLNL DDFFSFLFSD DLNHPIDSKV HQDMASPLSH 120
121 YFIYTSHNSY LTGNQINSEC SDVPLIKALK RGVRALELDM WPNSTKDDIL VLHGWAWTPP 180
181 VELVKCLRSI KEHAFYASAY PVILTLEDHL TPDLQAKAAE MMKEIFMDMV YFPEAGGLKE 240
241 FPSPEDLKYK IVISTKPPKG SLRKDKDSES DASGKASSDV SADDEKTEEE TSEAKNEEDG 300
301 FDQESSNLDF LTYSRLITIP SGNAKNGLKE ALTIDNGGVR RLSLREQKFK KATEMYGTEV 360
361 IKFTQKNLLR IYPKATRVNS SNYRPYNGWM YGAQMVAFNM QGYGRALWMM HGMFRGNGGC 420
421 GYVKKPDFMM NNNLSGEVFN PKAKLPIKKT LKVKVYMGKG WDSGFQRTCF NTWSSPNFYT 480
481 RVGITGVRGD KVMKKTKKEQ KTWEPFWNEE FEFQLTVPEL ALLRIEVHDY NMPEKDDFSG 540
541 QTCLPVSELR QGIRSVPLYD RKGERLVSVT LLMRFHFL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
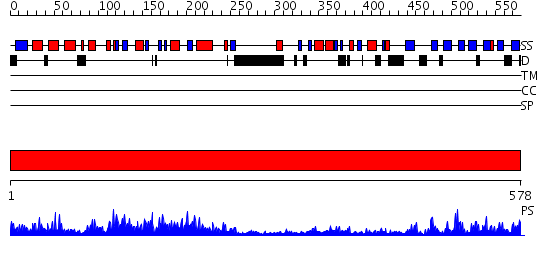
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..578] | 164.0 | Phosphoinositide-specific phospholipase C, isozyme D1 (PLC-D!); PI-specific phospholipase C isozyme D1 (PLC-D1), C-terminal domain; Phospholipase C isozyme D1 (PLC-D1) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)