













| General Information: |
|
| Name(s) found: |
ERCC2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 758 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
DNA repair
[IMP]
response to heat [IMP] response to UV [IMP] |
| Molecular Function: |
nucleic acid binding
[IEA]
DNA binding [IEA] ATP binding [IEA] hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides [IEA] ATP-dependent helicase activity [IEA] ATP-dependent DNA helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIFKIEDVTV YFPYDNIYPE QYEYMVELKR ALDAKGHCLL EMPTGTGKTI ALLSLITSYR 60
61 LSRPDSPIKL VYCTRTVHEM EKTLGELKLL HDYQVRHLGT QAKILALGLS SRKNLCVNTK 120
121 VLAAENRDSV DAACRKRTAS WVRALSTENP NVELCDFFEN YEKAAENALL PPGVYTLEDL 180
181 RAFGKNRGWC PYFLARHMIQ FANVIVYSYQ YLLDPKVAGF ISKELQKESV VVFDEAHNID 240
241 NVCIEALSVS VRRVTLEGAN RNLNKIRQEI DRFKATDAGR LRAEYNRLVE GLALRGDLSG 300
301 GDQWLANPAL PHDILKEAVP GNIRRAEHFV HVLRRLLQYL GVRLDTENVE KESPVSFVSS 360
361 LNSQAGIEQK TLKFCYDRLQ SLMLTLEITD TDEFLPIQTV CDFATLVGTY ARGFSIIIEP 420
421 YDERMPHIPD PILQLSCHDA SLAIKPVFDR FQSVVITSGT LSPIDLYPRL LNFTPVVSRS 480
481 FKMSMTRDCI CPMVLTRGSD QLPVSTKFDM RSDPGVVRNY GKLLVEMVSI VPDGVVCFFV 540
541 SYSYMDGIIA TWNETGILKE IMQQKLVFIE TQDVVETTLA LDNYRRACDC GRGAVFFSVA 600
601 RGKVAEGIDF DRHYGRLVVM YGVPFQYTLS KILRARLEYL HDTFQIKEGD FLTFDALRQA 660
661 AQCVGRVIRS KADYGMMIFA DKRYSRHDKR SKLPGWILSH LRDAHLNLST DMAIHIAREF 720
721 LRKMAQPYDK AGTMGRKTLL TQEDLEKMAE TGVQDMAY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
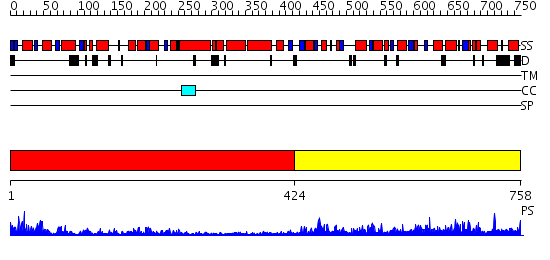
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..423] | 12.30103 | No description for 2i4iA was found. | |
| 2 | View Details | [424..758] | 0.984 | No description for 2ocaA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)